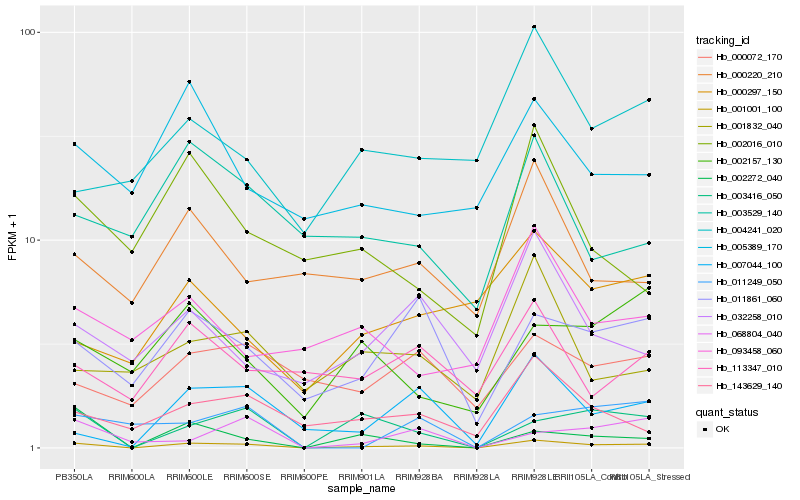
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001001_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650392 [Jatropha curcas] |
| 2 |
Hb_003416_050 |
0.2439989284 |
- |
- |
- |
| 3 |
Hb_002272_040 |
0.2694883699 |
- |
- |
hypothetical protein POPTR_0013s02610g, partial [Populus trichocarpa] |
| 4 |
Hb_007044_100 |
0.2699385612 |
- |
- |
hypothetical protein POPTR_0004s10420g [Populus trichocarpa] |
| 5 |
Hb_011861_060 |
0.2850270305 |
- |
- |
- |
| 6 |
Hb_143629_140 |
0.2973607181 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 7 |
Hb_002157_130 |
0.3049204928 |
- |
- |
- |
| 8 |
Hb_068804_040 |
0.3074536818 |
- |
- |
- |
| 9 |
Hb_002016_010 |
0.3142403449 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011249_050 |
0.3195223999 |
- |
- |
- |
| 11 |
Hb_005389_170 |
0.3205469919 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 12 |
Hb_000220_210 |
0.320595118 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000072_170 |
0.3206870021 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 14 |
Hb_032258_010 |
0.3225143898 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 15 |
Hb_003529_140 |
0.3226770467 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_113347_010 |
0.323083049 |
- |
- |
hypothetical protein CISIN_1g009336mg [Citrus sinensis] |
| 17 |
Hb_000297_150 |
0.3243654172 |
- |
- |
PREDICTED: uncharacterized protein LOC103500908 [Cucumis melo] |
| 18 |
Hb_001832_040 |
0.3258829524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_093458_060 |
0.3260970866 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 20 |
Hb_004241_020 |
0.326523664 |
- |
- |
PREDICTED: DAG protein, chloroplastic-like [Jatropha curcas] |