| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
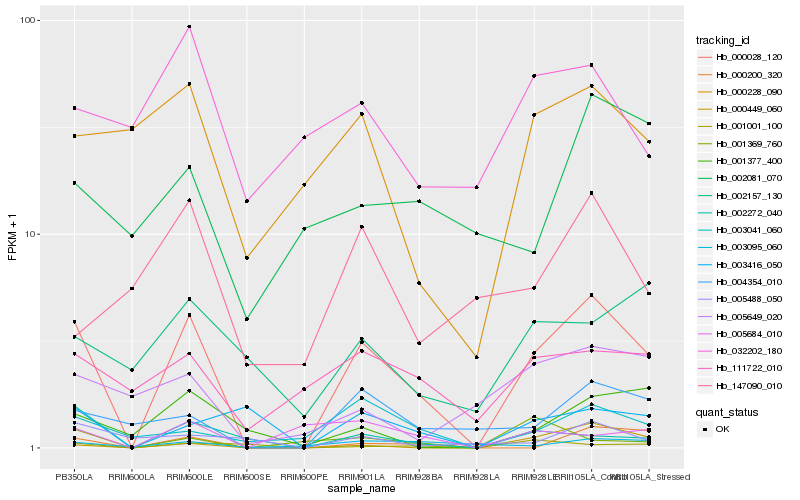
Hb_000028_120 |
0.0 |
- |
- |
Os03g0221800 [Oryza sativa Japonica Group] |
| 2 |
Hb_002272_040 |
0.2986557416 |
- |
- |
hypothetical protein POPTR_0013s02610g, partial [Populus trichocarpa] |
| 3 |
Hb_004354_010 |
0.2991440855 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 4 |
Hb_000200_320 |
0.3032376929 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein JCGZ_09311 [Jatropha curcas] |
| 5 |
Hb_003041_060 |
0.3118853818 |
- |
- |
- |
| 6 |
Hb_111722_010 |
0.3263558412 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X2 [Jatropha curcas] |
| 7 |
Hb_003416_050 |
0.3295623705 |
- |
- |
- |
| 8 |
Hb_001377_400 |
0.3330117036 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 9 |
Hb_003095_060 |
0.3330215735 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 10 |
Hb_000228_090 |
0.3394272317 |
- |
- |
PREDICTED: protein PAM68, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005649_020 |
0.3450583411 |
- |
- |
PREDICTED: serine carboxypeptidase-like 25 [Jatropha curcas] |
| 12 |
Hb_005684_010 |
0.3456290176 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 13 |
Hb_000449_060 |
0.3473483589 |
- |
- |
- |
| 14 |
Hb_002157_130 |
0.349619096 |
- |
- |
- |
| 15 |
Hb_001001_100 |
0.3540489928 |
- |
- |
PREDICTED: uncharacterized protein LOC105650392 [Jatropha curcas] |
| 16 |
Hb_002081_070 |
0.3614066919 |
- |
- |
PREDICTED: pyridoxal kinase [Jatropha curcas] |
| 17 |
Hb_005488_050 |
0.3638777927 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: retrovirus-related Pol polyprotein from transposon TNT 1-94 [Cucumis sativus] |
| 18 |
Hb_001369_760 |
0.3653989802 |
- |
- |
hypothetical protein PRUPE_ppa019417mg [Prunus persica] |
| 19 |
Hb_032202_180 |
0.3656899024 |
- |
- |
PREDICTED: protein-ribulosamine 3-kinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_147090_010 |
0.3662309783 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Malus domestica] |