| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
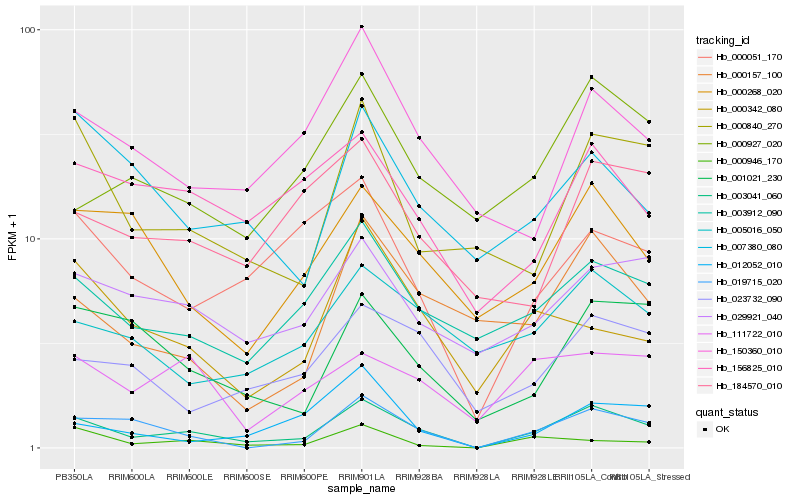
Hb_003041_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_019715_020 |
0.1731803304 |
- |
- |
- |
| 3 |
Hb_003912_090 |
0.1931550781 |
- |
- |
peptidyl-tRNA hydrolase, putative [Ricinus communis] |
| 4 |
Hb_001021_230 |
0.2031374196 |
- |
- |
- |
| 5 |
Hb_000840_270 |
0.2031613151 |
- |
- |
PREDICTED: small RNA degrading nuclease 1 [Jatropha curcas] |
| 6 |
Hb_029921_040 |
0.2071563938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_023732_090 |
0.2090648386 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_000268_020 |
0.2096203001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000157_100 |
0.2099900696 |
- |
- |
hypothetical protein B456_007G083500 [Gossypium raimondii] |
| 10 |
Hb_000051_170 |
0.2112821726 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_150360_010 |
0.2115566696 |
- |
- |
hypothetical protein JCGZ_16890 [Jatropha curcas] |
| 12 |
Hb_005016_050 |
0.2164964224 |
- |
- |
- |
| 13 |
Hb_111722_010 |
0.2180857751 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000927_020 |
0.2194015025 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 15 |
Hb_156825_010 |
0.2195702225 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |
| 16 |
Hb_000342_080 |
0.2223664714 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 17 |
Hb_007380_080 |
0.2237548189 |
transcription factor |
TF Family: G2-like |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 18 |
Hb_184570_010 |
0.2239592972 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 19 |
Hb_012052_010 |
0.2243678449 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 20 |
Hb_000946_170 |
0.2262416569 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |