| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
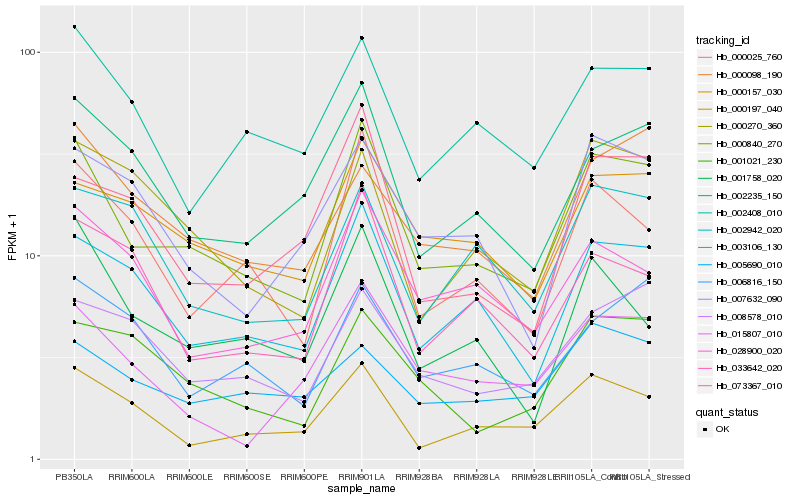
Hb_000840_270 |
0.0 |
- |
- |
PREDICTED: small RNA degrading nuclease 1 [Jatropha curcas] |
| 2 |
Hb_003106_130 |
0.1138866264 |
- |
- |
PREDICTED: uncharacterized protein LOC105640436 [Jatropha curcas] |
| 3 |
Hb_000157_030 |
0.1281467142 |
- |
- |
PREDICTED: cytochrome b5 domain-containing protein RLF [Jatropha curcas] |
| 4 |
Hb_000025_760 |
0.131533109 |
- |
- |
- |
| 5 |
Hb_002235_150 |
0.1323796204 |
- |
- |
- |
| 6 |
Hb_002408_010 |
0.138595403 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 7 |
Hb_008578_010 |
0.1402206154 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 8 |
Hb_000197_040 |
0.1408497986 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 9 |
Hb_000270_360 |
0.1439270403 |
- |
- |
PREDICTED: serine/threonine-protein kinase pakF-like [Jatropha curcas] |
| 10 |
Hb_073367_010 |
0.1460049762 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |
| 11 |
Hb_015807_010 |
0.1464689405 |
- |
- |
hypothetical protein POPTR_0001s44430g [Populus trichocarpa] |
| 12 |
Hb_000098_190 |
0.1466144802 |
- |
- |
PREDICTED: uncharacterized protein LOC105633343 [Jatropha curcas] |
| 13 |
Hb_028900_020 |
0.1468133247 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_007632_090 |
0.1480878739 |
- |
- |
PREDICTED: nicotinamide/nicotinic acid mononucleotide adenylyltransferase isoform X1 [Jatropha curcas] |
| 15 |
Hb_006816_150 |
0.1497023762 |
- |
- |
PREDICTED: lon protease homolog 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001758_020 |
0.1506862319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001021_230 |
0.1506904753 |
- |
- |
- |
| 18 |
Hb_005690_010 |
0.1518173982 |
- |
- |
PREDICTED: origin recognition complex subunit 2 [Populus euphratica] |
| 19 |
Hb_002942_020 |
0.1545727716 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 20 |
Hb_033642_020 |
0.154837629 |
- |
- |
hypothetical protein JCGZ_19094 [Jatropha curcas] |