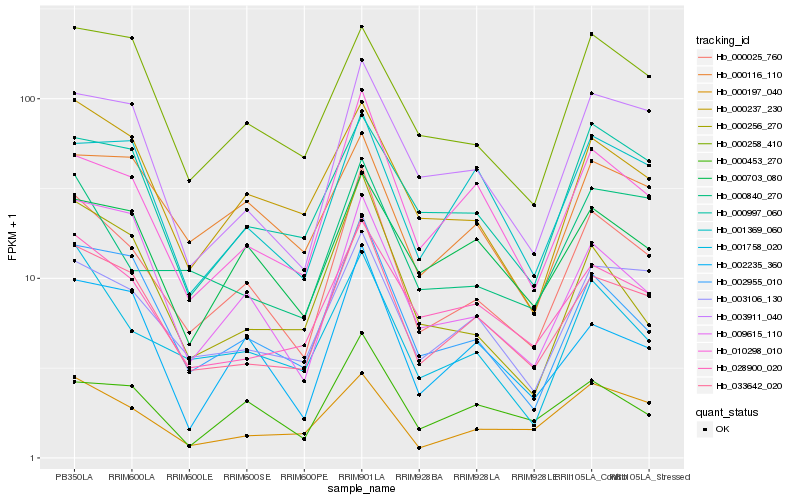
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_760 |
0.0 |
- |
- |
- |
| 2 |
Hb_033642_020 |
0.0952127553 |
- |
- |
hypothetical protein JCGZ_19094 [Jatropha curcas] |
| 3 |
Hb_003106_130 |
0.112683341 |
- |
- |
PREDICTED: uncharacterized protein LOC105640436 [Jatropha curcas] |
| 4 |
Hb_002955_010 |
0.1131816343 |
- |
- |
PREDICTED: Werner syndrome ATP-dependent helicase homolog [Jatropha curcas] |
| 5 |
Hb_001758_020 |
0.1155893138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010298_010 |
0.1167502207 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 [Jatropha curcas] |
| 7 |
Hb_003911_040 |
0.1218376527 |
- |
- |
PREDICTED: putative H/ACA ribonucleoprotein complex subunit 1-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000237_230 |
0.1235367893 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000197_040 |
0.1258167096 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_009615_110 |
0.1276364224 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 11 |
Hb_028900_020 |
0.1292632799 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000840_270 |
0.131533109 |
- |
- |
PREDICTED: small RNA degrading nuclease 1 [Jatropha curcas] |
| 13 |
Hb_000258_410 |
0.1328927433 |
- |
- |
PREDICTED: uncharacterized protein LOC105630409 [Jatropha curcas] |
| 14 |
Hb_002235_360 |
0.1367465137 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 15 |
Hb_000997_060 |
0.1369225671 |
- |
- |
PREDICTED: DNA repair protein XRCC4 [Jatropha curcas] |
| 16 |
Hb_000116_110 |
0.138034024 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 17 |
Hb_000453_270 |
0.1387894573 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 [Propithecus coquereli] |
| 18 |
Hb_000256_270 |
0.1400671752 |
- |
- |
PREDICTED: large proline-rich protein bag6-B isoform X1 [Jatropha curcas] |
| 19 |
Hb_000703_080 |
0.1407318997 |
- |
- |
guanine nucleotide exchange factor P532, putative [Ricinus communis] |
| 20 |
Hb_001369_060 |
0.1435506863 |
- |
- |
PREDICTED: uncharacterized Rho GTPase-activating protein At5g61530 [Jatropha curcas] |