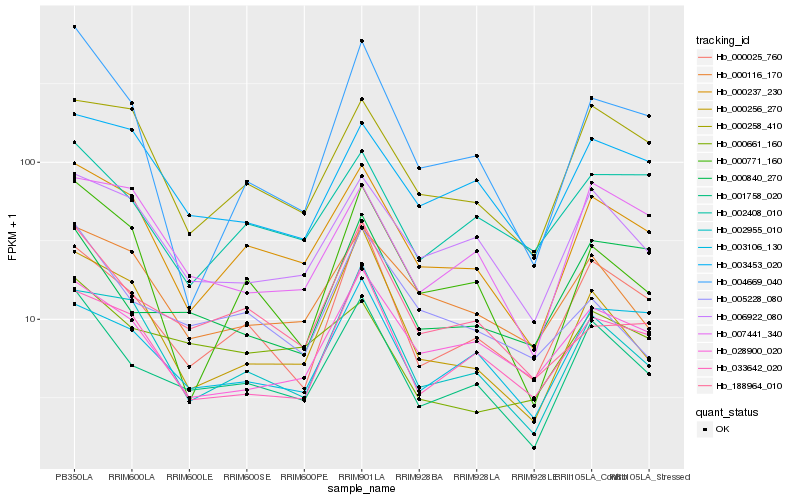
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001758_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000025_760 |
0.1155893138 |
- |
- |
- |
| 3 |
Hb_000237_230 |
0.1209621836 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006922_080 |
0.135982781 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT3-like [Glycine max] |
| 5 |
Hb_002955_010 |
0.1482278517 |
- |
- |
PREDICTED: Werner syndrome ATP-dependent helicase homolog [Jatropha curcas] |
| 6 |
Hb_000840_270 |
0.1506862319 |
- |
- |
PREDICTED: small RNA degrading nuclease 1 [Jatropha curcas] |
| 7 |
Hb_005228_080 |
0.1507391584 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 8 |
Hb_028900_020 |
0.1507981886 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000256_270 |
0.1511226664 |
- |
- |
PREDICTED: large proline-rich protein bag6-B isoform X1 [Jatropha curcas] |
| 10 |
Hb_033642_020 |
0.1517093915 |
- |
- |
hypothetical protein JCGZ_19094 [Jatropha curcas] |
| 11 |
Hb_003106_130 |
0.1566655626 |
- |
- |
PREDICTED: uncharacterized protein LOC105640436 [Jatropha curcas] |
| 12 |
Hb_000116_170 |
0.1579349683 |
- |
- |
hypothetical protein JCGZ_22392 [Jatropha curcas] |
| 13 |
Hb_188964_010 |
0.1584355532 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 14 |
Hb_004669_040 |
0.1590321689 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 15 |
Hb_002408_010 |
0.1616386303 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 16 |
Hb_000258_410 |
0.1620809255 |
- |
- |
PREDICTED: uncharacterized protein LOC105630409 [Jatropha curcas] |
| 17 |
Hb_000771_160 |
0.1624107961 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 18 |
Hb_007441_340 |
0.1625375766 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_003453_020 |
0.1633544251 |
- |
- |
PREDICTED: histidine biosynthesis bifunctional protein hisIE, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000661_160 |
0.1633715438 |
- |
- |
hexokinase [Manihot esculenta] |