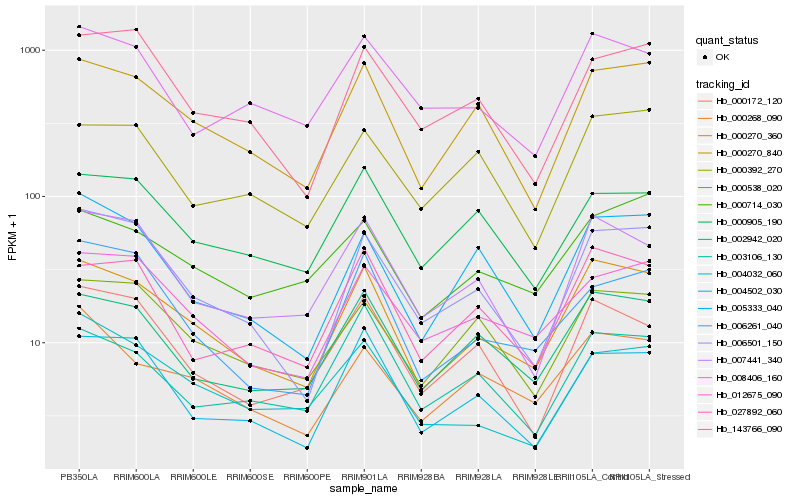
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_360 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase pakF-like [Jatropha curcas] |
| 2 |
Hb_000270_840 |
0.0844544954 |
- |
- |
PREDICTED: probable 26S proteasome complex subunit sem1-2 [Nicotiana tomentosiformis] |
| 3 |
Hb_002942_020 |
0.0883822373 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 4 |
Hb_006501_150 |
0.1000881061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_027892_060 |
0.1032868246 |
- |
- |
PREDICTED: inter-alpha-trypsin inhibitor heavy chain H3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007441_340 |
0.1054904265 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_012675_090 |
0.1092326791 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_000268_090 |
0.1095396048 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004502_030 |
0.1103019941 |
- |
- |
unknown [Populus trichocarpa] |
| 10 |
Hb_000538_020 |
0.1107189662 |
- |
- |
PREDICTED: autophagy protein 5 [Jatropha curcas] |
| 11 |
Hb_005333_040 |
0.1141319483 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 5 [Prunus mume] |
| 12 |
Hb_000172_120 |
0.1168778864 |
- |
- |
PREDICTED: uncharacterized protein LOC105650770 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000714_030 |
0.1170130528 |
- |
- |
Nucleosome assembly [Gossypium arboreum] |
| 14 |
Hb_008406_160 |
0.1218136051 |
- |
- |
PREDICTED: ADP,ATP carrier protein 3, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000905_190 |
0.1246786518 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm7 [Elaeis guineensis] |
| 16 |
Hb_004032_060 |
0.1251033213 |
- |
- |
elongation factor 1 gamma-like protein, partial [Ipomoea nil] |
| 17 |
Hb_003106_130 |
0.1252887798 |
- |
- |
PREDICTED: uncharacterized protein LOC105640436 [Jatropha curcas] |
| 18 |
Hb_006261_040 |
0.1254710675 |
- |
- |
PREDICTED: uncharacterized protein LOC105646602 [Jatropha curcas] |
| 19 |
Hb_143766_090 |
0.1262609219 |
- |
- |
60S ribosomal protein L31B [Hevea brasiliensis] |
| 20 |
Hb_000392_270 |
0.1266673446 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |