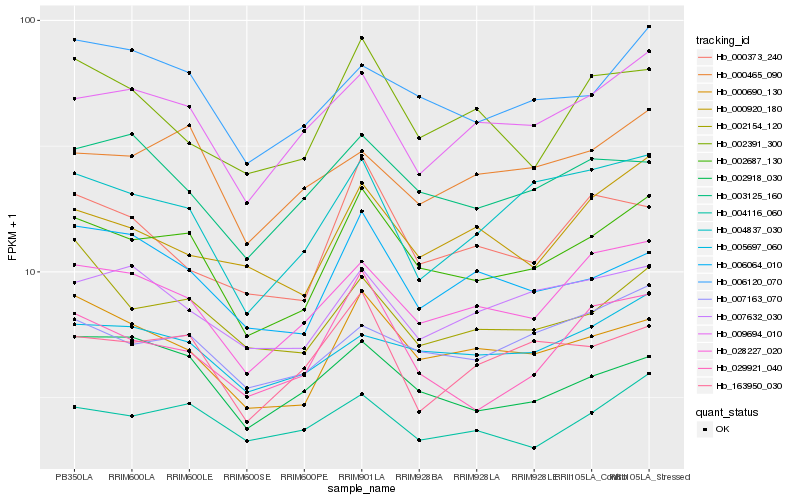
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631153 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000690_130 |
0.0703898513 |
- |
- |
PREDICTED: uncharacterized protein LOC105631771 [Jatropha curcas] |
| 3 |
Hb_028227_020 |
0.0775284267 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB isoform X1 [Jatropha curcas] |
| 4 |
Hb_006120_070 |
0.0831496738 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 5 |
Hb_009694_010 |
0.0836811794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_029921_040 |
0.0880867636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006064_010 |
0.0893486747 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 4 [Jatropha curcas] |
| 8 |
Hb_007163_070 |
0.0904682966 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_005697_060 |
0.0914614995 |
transcription factor |
TF Family: HB |
PREDICTED: pathogenesis-related homeodomain protein isoform X1 [Jatropha curcas] |
| 10 |
Hb_004837_030 |
0.0915696126 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 [Jatropha curcas] |
| 11 |
Hb_004116_060 |
0.091687605 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 12 |
Hb_163950_030 |
0.093584985 |
- |
- |
PREDICTED: uncharacterized protein LOC105648509 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002918_030 |
0.0941117331 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 14 |
Hb_007632_030 |
0.0947456641 |
- |
- |
PREDICTED: poly(A)-specific ribonuclease PARN isoform X1 [Jatropha curcas] |
| 15 |
Hb_002391_300 |
0.0950852484 |
- |
- |
hypothetical protein PHAVU_002G104100g [Phaseolus vulgaris] |
| 16 |
Hb_000373_240 |
0.0957758685 |
- |
- |
PREDICTED: uncharacterized protein LOC105640940 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002154_120 |
0.0964267572 |
- |
- |
PREDICTED: uncharacterized protein LOC105630563 [Jatropha curcas] |
| 18 |
Hb_003125_160 |
0.0979772824 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000920_180 |
0.0980213772 |
- |
- |
PREDICTED: F-box protein SKIP5 [Jatropha curcas] |
| 20 |
Hb_000465_090 |
0.0989367905 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |