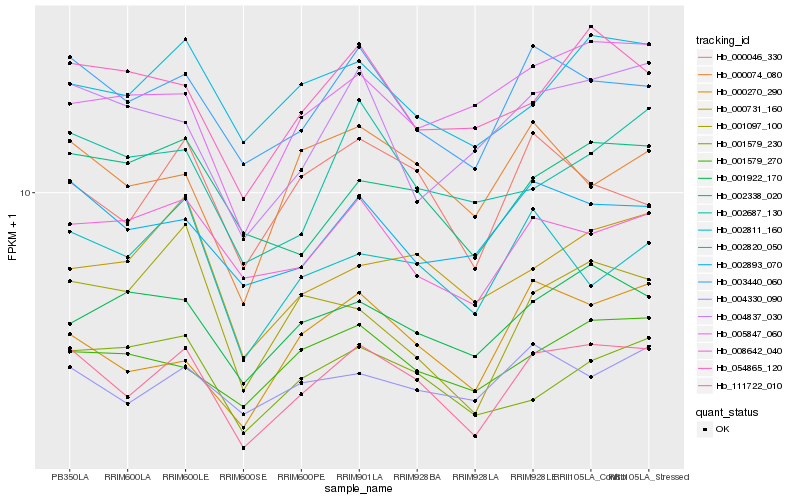
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111722_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003440_060 |
0.1235929017 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 3 |
Hb_000270_290 |
0.1316133595 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 4 |
Hb_000046_330 |
0.1397396475 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 5 |
Hb_002811_160 |
0.1427820046 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000074_080 |
0.1430440398 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_004330_090 |
0.1431113713 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_002893_070 |
0.1437354508 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001579_270 |
0.143796652 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 10 |
Hb_002338_020 |
0.1440003163 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 11 |
Hb_004837_030 |
0.1440160374 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 [Jatropha curcas] |
| 12 |
Hb_054865_120 |
0.1449476448 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 13 |
Hb_001579_230 |
0.145397582 |
- |
- |
PREDICTED: uncharacterized protein LOC105644968 [Jatropha curcas] |
| 14 |
Hb_000731_160 |
0.1456534377 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 15 |
Hb_001922_170 |
0.1459229567 |
- |
- |
Plastidic pyruvate kinase beta subunit 1 isoform 2 [Theobroma cacao] |
| 16 |
Hb_001097_100 |
0.1471066971 |
- |
- |
Chaperone DnaJ-domain superfamily protein [Theobroma cacao] |
| 17 |
Hb_002820_050 |
0.1473877697 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 18 |
Hb_008642_040 |
0.1479044712 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002687_130 |
0.1481992576 |
- |
- |
PREDICTED: uncharacterized protein LOC105631153 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005847_060 |
0.1496408811 |
- |
- |
hypothetical protein POPTR_0006s02920g [Populus trichocarpa] |