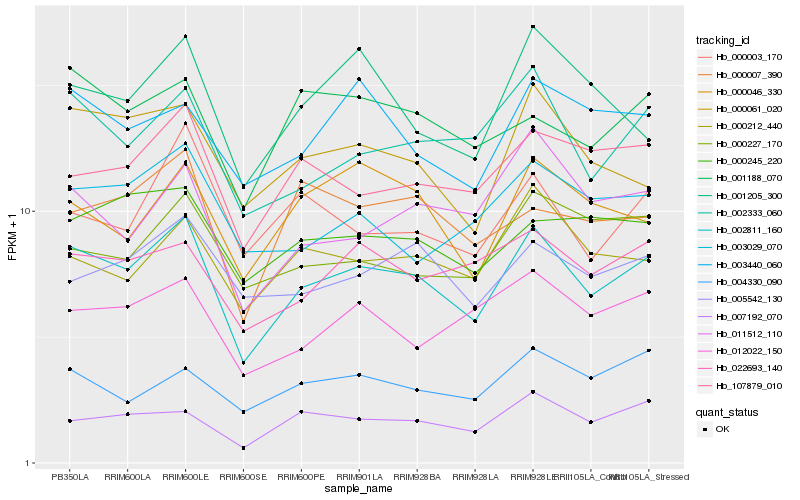
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_160 |
0.0 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002333_060 |
0.0940285938 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_107879_010 |
0.1008040121 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 4 |
Hb_000061_020 |
0.1010808647 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 5 |
Hb_007192_070 |
0.1019312993 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_022693_140 |
0.1036884712 |
- |
- |
PREDICTED: uncharacterized protein LOC105632654 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000212_440 |
0.1054995299 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001188_070 |
0.10584634 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 [Jatropha curcas] |
| 9 |
Hb_012022_150 |
0.1067065938 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 10 |
Hb_005542_130 |
0.1094072837 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 11 |
Hb_000245_220 |
0.1094139372 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 12 |
Hb_000046_330 |
0.1100989828 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 13 |
Hb_011512_110 |
0.1101010379 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_004330_090 |
0.1104590253 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_003440_060 |
0.1105877165 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 16 |
Hb_003029_070 |
0.1109325367 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000007_390 |
0.1113970536 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 18 |
Hb_000227_170 |
0.1127843761 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_000003_170 |
0.1128142772 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 20 |
Hb_001205_300 |
0.113625743 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |