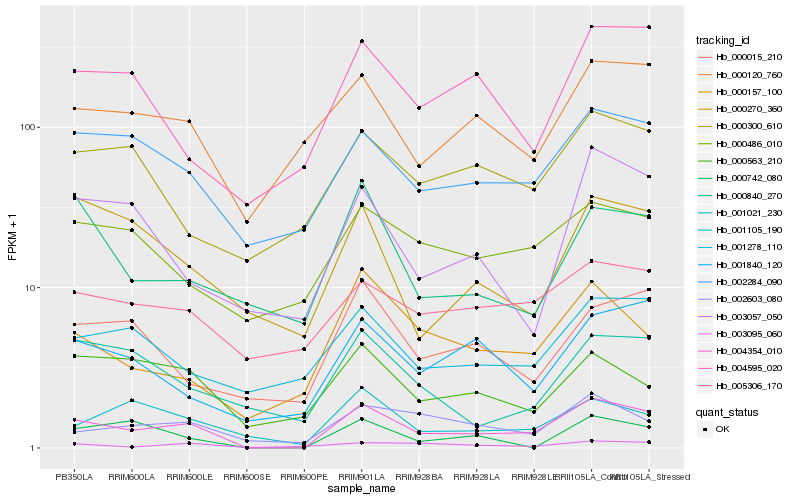
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004354_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 2 |
Hb_002284_090 |
0.2064498872 |
- |
- |
PREDICTED: outer envelope pore protein 21B, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000157_100 |
0.2144537406 |
- |
- |
hypothetical protein B456_007G083500 [Gossypium raimondii] |
| 4 |
Hb_002603_080 |
0.2177599869 |
- |
- |
hypothetical protein MTR_7g114400 [Medicago truncatula] |
| 5 |
Hb_001840_120 |
0.2184881432 |
- |
- |
PREDICTED: uncharacterized protein LOC104228746 [Nicotiana sylvestris] |
| 6 |
Hb_000120_760 |
0.2197584069 |
transcription factor |
TF Family: HMG |
PREDICTED: HMG1/2-like protein [Populus euphratica] |
| 7 |
Hb_004595_020 |
0.2204119031 |
- |
- |
PREDICTED: probable prefoldin subunit 5 [Jatropha curcas] |
| 8 |
Hb_001105_190 |
0.2208890871 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 9 |
Hb_003057_050 |
0.2226477385 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 10 |
Hb_001278_110 |
0.2251344832 |
- |
- |
PREDICTED: uncharacterized protein LOC104613296 isoform X3 [Nelumbo nucifera] |
| 11 |
Hb_000015_210 |
0.2253958063 |
- |
- |
PREDICTED: uncharacterized protein LOC105647781 [Jatropha curcas] |
| 12 |
Hb_003095_060 |
0.2289746902 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 13 |
Hb_001021_230 |
0.2295740232 |
- |
- |
- |
| 14 |
Hb_000270_360 |
0.2296533332 |
- |
- |
PREDICTED: serine/threonine-protein kinase pakF-like [Jatropha curcas] |
| 15 |
Hb_005306_170 |
0.2301234359 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 16 |
Hb_000840_270 |
0.2305798947 |
- |
- |
PREDICTED: small RNA degrading nuclease 1 [Jatropha curcas] |
| 17 |
Hb_000563_210 |
0.2315450439 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 20a [Jatropha curcas] |
| 18 |
Hb_000486_010 |
0.2318647221 |
- |
- |
PREDICTED: uncharacterized protein LOC105646661 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000742_080 |
0.232671027 |
- |
- |
- |
| 20 |
Hb_000300_610 |
0.2327163285 |
- |
- |
cytosolic copper/zinc superoxide dismutase [Hevea brasiliensis] |