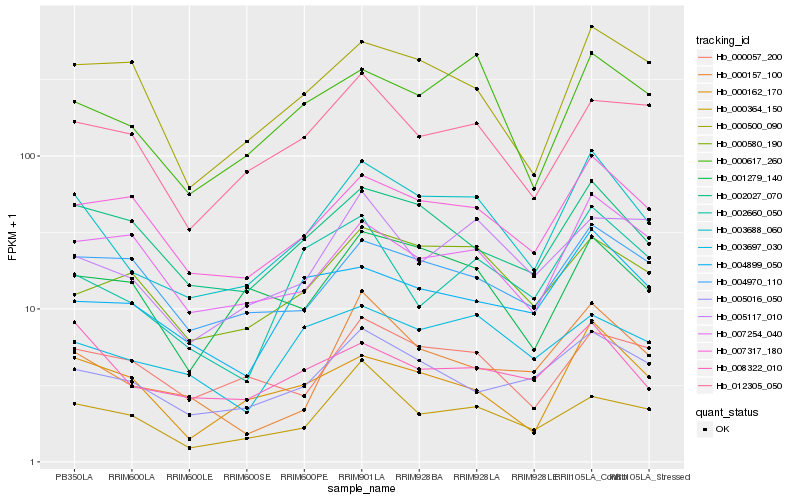
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_060 |
0.0 |
- |
- |
PREDICTED: histone deacetylase 5 [Jatropha curcas] |
| 2 |
Hb_000617_260 |
0.1355670482 |
rubber biosynthesis |
Gene Name: SRPP2 |
small rubber particle protein [Hevea brasiliensis] |
| 3 |
Hb_007317_180 |
0.141142386 |
- |
- |
hypothetical protein CICLE_v10028933mg [Citrus clementina] |
| 4 |
Hb_002027_070 |
0.1449290864 |
- |
- |
Auxin-binding protein T85 precursor, putative [Ricinus communis] |
| 5 |
Hb_001279_140 |
0.1471524945 |
- |
- |
PREDICTED: cystinosin homolog [Jatropha curcas] |
| 6 |
Hb_000157_100 |
0.147463757 |
- |
- |
hypothetical protein B456_007G083500 [Gossypium raimondii] |
| 7 |
Hb_005016_050 |
0.1480045336 |
- |
- |
- |
| 8 |
Hb_000580_190 |
0.1510556186 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 9 |
Hb_000162_170 |
0.1536557029 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 10 |
Hb_000057_200 |
0.1571268529 |
- |
- |
PREDICTED: methyltransferase-like protein 22 [Jatropha curcas] |
| 11 |
Hb_008322_010 |
0.1586233328 |
- |
- |
PREDICTED: DNA repair protein RAD51 homolog 3 [Jatropha curcas] |
| 12 |
Hb_004970_110 |
0.1588579747 |
- |
- |
PREDICTED: protein ROOT INITIATION DEFECTIVE 3 [Jatropha curcas] |
| 13 |
Hb_000364_150 |
0.1594138157 |
- |
- |
- |
| 14 |
Hb_002660_050 |
0.1604689111 |
- |
- |
PREDICTED: uncharacterized protein LOC105642218 [Jatropha curcas] |
| 15 |
Hb_000500_090 |
0.1604712575 |
- |
- |
nucleoside diphosphate kinase 2 [Hevea brasiliensis] |
| 16 |
Hb_004899_050 |
0.1607902104 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 17 |
Hb_003697_030 |
0.1608524382 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATXR2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012305_050 |
0.1609044898 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_005117_010 |
0.161148482 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_007254_040 |
0.1629727175 |
- |
- |
PREDICTED: protein MITOFERRINLIKE 1, chloroplastic [Jatropha curcas] |