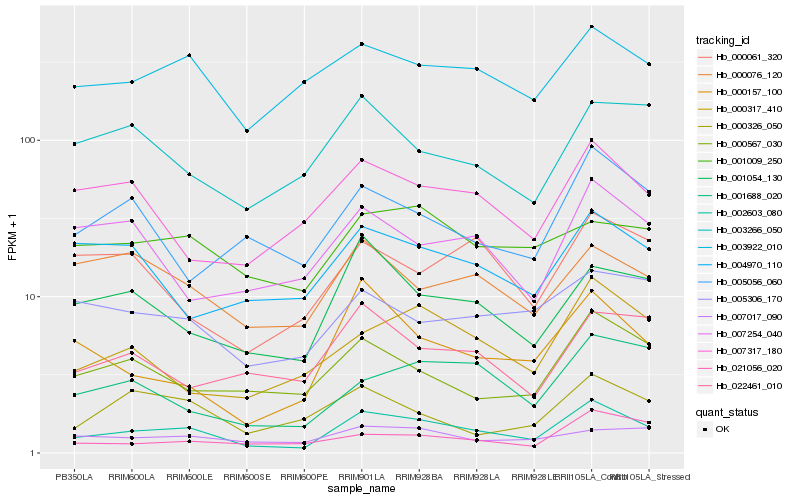
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002603_080 |
0.0 |
- |
- |
hypothetical protein MTR_7g114400 [Medicago truncatula] |
| 2 |
Hb_001054_130 |
0.1594877445 |
- |
- |
Protein SSU72, putative [Ricinus communis] |
| 3 |
Hb_003922_010 |
0.1638636446 |
- |
- |
PREDICTED: probable phospholipid hydroperoxide glutathione peroxidase [Jatropha curcas] |
| 4 |
Hb_000567_030 |
0.1656736885 |
- |
- |
hypothetical protein JCGZ_21550 [Jatropha curcas] |
| 5 |
Hb_000317_410 |
0.1657744916 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 6 |
Hb_000157_100 |
0.1679591644 |
- |
- |
hypothetical protein B456_007G083500 [Gossypium raimondii] |
| 7 |
Hb_007317_180 |
0.1730730292 |
- |
- |
hypothetical protein CICLE_v10028933mg [Citrus clementina] |
| 8 |
Hb_001688_020 |
0.175732515 |
- |
- |
PREDICTED: uncharacterized protein LOC103438026 [Malus domestica] |
| 9 |
Hb_022461_010 |
0.1785440516 |
- |
- |
- |
| 10 |
Hb_001009_250 |
0.1786749726 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005056_060 |
0.178867346 |
- |
- |
PREDICTED: prostamide/prostaglandin F synthase isoform X1 [Jatropha curcas] |
| 12 |
Hb_000076_120 |
0.1813905855 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 13 |
Hb_004970_110 |
0.1815951 |
- |
- |
PREDICTED: protein ROOT INITIATION DEFECTIVE 3 [Jatropha curcas] |
| 14 |
Hb_005306_170 |
0.1824204341 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 15 |
Hb_007254_040 |
0.1833500497 |
- |
- |
PREDICTED: protein MITOFERRINLIKE 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_021056_020 |
0.1843589916 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 17 |
Hb_003266_050 |
0.1848482648 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D1 [Amborella trichopoda] |
| 18 |
Hb_000326_050 |
0.1849320925 |
- |
- |
Phenylalanyl-tRNA synthetase alpha chain isoform 2 [Theobroma cacao] |
| 19 |
Hb_000061_320 |
0.1865432689 |
- |
- |
PREDICTED: uncharacterized protein LOC105640575 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007017_090 |
0.1865456809 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 186 [Jatropha curcas] |