| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
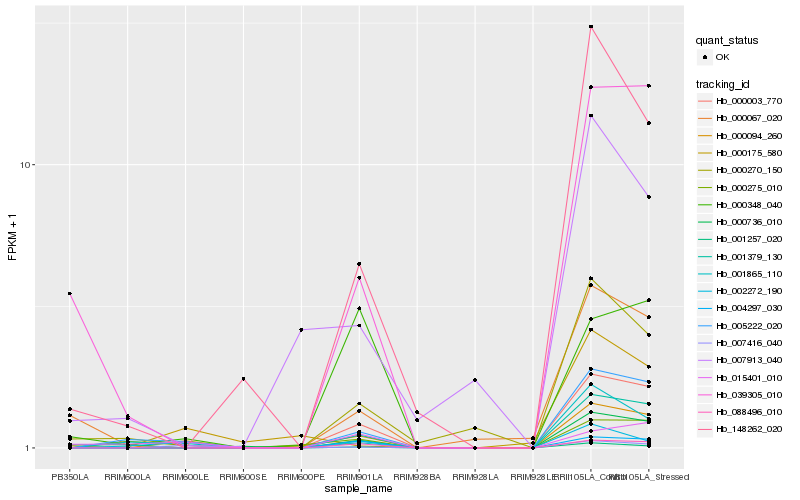
| 1 |
Hb_000094_260 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007416_040 |
0.2084384608 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 3 |
Hb_005222_020 |
0.2128736797 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000003_770 |
0.2279651315 |
- |
- |
- |
| 5 |
Hb_001379_130 |
0.2291961757 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 6 |
Hb_039305_010 |
0.2454441431 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 7 |
Hb_000275_010 |
0.2467537498 |
- |
- |
- |
| 8 |
Hb_001257_020 |
0.2512634014 |
- |
- |
- |
| 9 |
Hb_000270_150 |
0.2557627069 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_088496_010 |
0.2557730954 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_000067_020 |
0.2601729732 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 12 |
Hb_148262_020 |
0.2610020982 |
- |
- |
- |
| 13 |
Hb_001865_110 |
0.2625391281 |
- |
- |
- |
| 14 |
Hb_000736_010 |
0.2671381779 |
- |
- |
PREDICTED: uncharacterized protein LOC105762083 [Gossypium raimondii] |
| 15 |
Hb_004297_030 |
0.2699898704 |
- |
- |
PREDICTED: uncharacterized protein LOC105633198 [Jatropha curcas] |
| 16 |
Hb_000175_580 |
0.2707126048 |
transcription factor, desease resistance |
TF Family: C3H, Gene Name: zf-CCCH |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 17 |
Hb_007913_040 |
0.2724887256 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 18 |
Hb_000348_040 |
0.2765166141 |
- |
- |
- |
| 19 |
Hb_002272_190 |
0.2768001714 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 20 |
Hb_015401_010 |
0.2774930436 |
- |
- |
hypothetical protein VITISV_001548 [Vitis vinifera] |