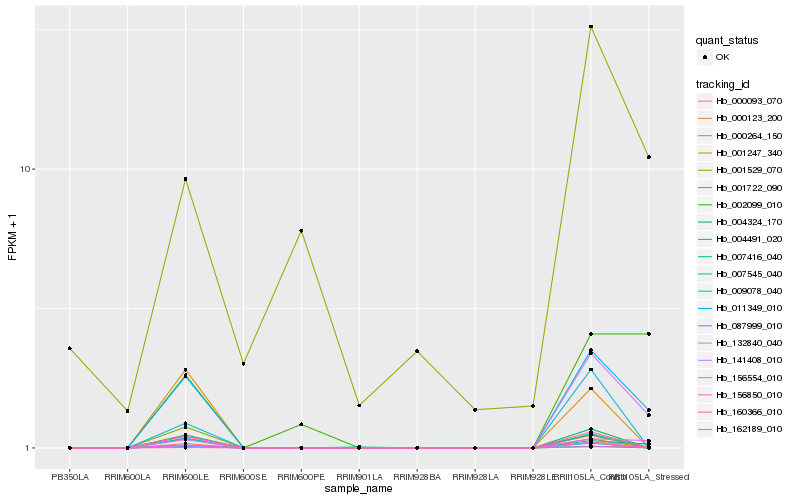
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011349_010 |
0.0 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 2 |
Hb_156554_010 |
0.1706533167 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 3 |
Hb_000123_200 |
0.2377789864 |
- |
- |
PREDICTED: WAT1-related protein At2g39510 [Jatropha curcas] |
| 4 |
Hb_160366_010 |
0.2443418085 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_007416_040 |
0.2509860847 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 6 |
Hb_004324_170 |
0.2832788369 |
- |
- |
- |
| 7 |
Hb_007545_040 |
0.283438864 |
- |
- |
PREDICTED: uncharacterized protein LOC104884077 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_162189_010 |
0.2836997739 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_002099_010 |
0.2859804052 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 10 |
Hb_156850_010 |
0.2873599277 |
- |
- |
PREDICTED: uncharacterized protein LOC105635403 [Jatropha curcas] |
| 11 |
Hb_001247_340 |
0.2964441629 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_132840_040 |
0.3074386981 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase, putative [Ricinus communis] |
| 13 |
Hb_000093_070 |
0.3110963237 |
transcription factor |
TF Family: C2C2-YABBY |
Protein CRABS CLAW, putative [Ricinus communis] |
| 14 |
Hb_087999_010 |
0.3113769387 |
- |
- |
PREDICTED: uncharacterized protein LOC105640118 [Jatropha curcas] |
| 15 |
Hb_004491_020 |
0.3210225386 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 16 |
Hb_000264_150 |
0.325150509 |
- |
- |
PREDICTED: uncharacterized protein LOC103870759 [Brassica rapa] |
| 17 |
Hb_141408_010 |
0.3301079859 |
- |
- |
PREDICTED: uncharacterized protein LOC105631382 [Jatropha curcas] |
| 18 |
Hb_009078_040 |
0.3331324145 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_001529_070 |
0.3393396952 |
transcription factor |
TF Family: AP2 |
hypothetical protein POPTR_0018s09900g [Populus trichocarpa] |
| 20 |
Hb_001722_090 |
0.3402672752 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |