| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
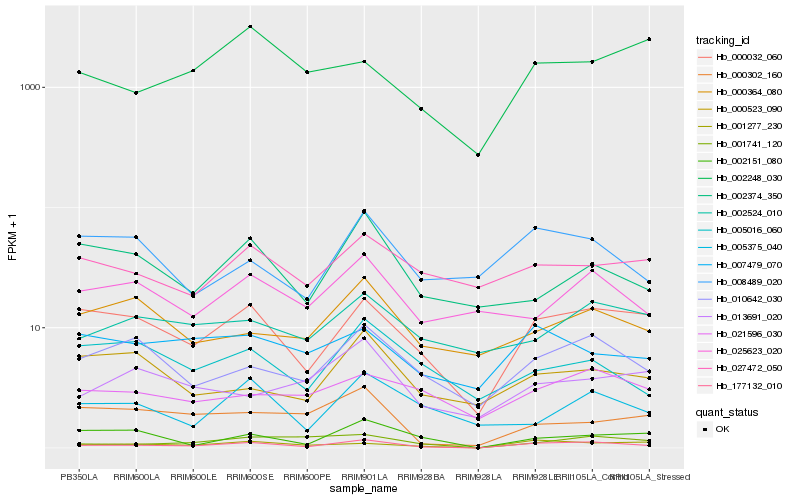
Hb_177132_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_000032_060 |
0.1672558394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001277_230 |
0.1941219621 |
- |
- |
- |
| 4 |
Hb_010642_030 |
0.2067361864 |
- |
- |
zinc phosphodiesterase, putative [Ricinus communis] |
| 5 |
Hb_001741_120 |
0.2195080386 |
- |
- |
hypothetical protein CARUB_v10011543mg, partial [Capsella rubella] |
| 6 |
Hb_008489_020 |
0.2347609934 |
- |
- |
PREDICTED: uncharacterized protein LOC105644663 [Jatropha curcas] |
| 7 |
Hb_002151_080 |
0.2352382561 |
- |
- |
hypothetical protein MIMGU_mgv1a000572mg [Erythranthe guttata] |
| 8 |
Hb_005375_040 |
0.2368083241 |
- |
- |
dicer-1, putative [Ricinus communis] |
| 9 |
Hb_002374_350 |
0.2384984112 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 10 |
Hb_025623_020 |
0.2387462522 |
- |
- |
PREDICTED: uncharacterized protein LOC105646971 [Jatropha curcas] |
| 11 |
Hb_002524_010 |
0.2431441521 |
- |
- |
hypothetical protein POPTR_0006s20250g [Populus trichocarpa] |
| 12 |
Hb_021596_030 |
0.2434154304 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007479_070 |
0.2436854269 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 14 |
Hb_005016_060 |
0.2446658637 |
- |
- |
PREDICTED: histidine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 15 |
Hb_000364_080 |
0.2447587001 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000302_160 |
0.2474276318 |
- |
- |
PREDICTED: uncharacterized protein LOC105648403 [Jatropha curcas] |
| 17 |
Hb_000523_090 |
0.2490108409 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 18 |
Hb_027472_050 |
0.2493224851 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 19 |
Hb_013691_020 |
0.2511777505 |
- |
- |
transcription initiation factor iie, alpha subunit, putative [Ricinus communis] |
| 20 |
Hb_002248_030 |
0.2515430028 |
- |
- |
metallothionein-like protein [Hevea brasiliensis] |