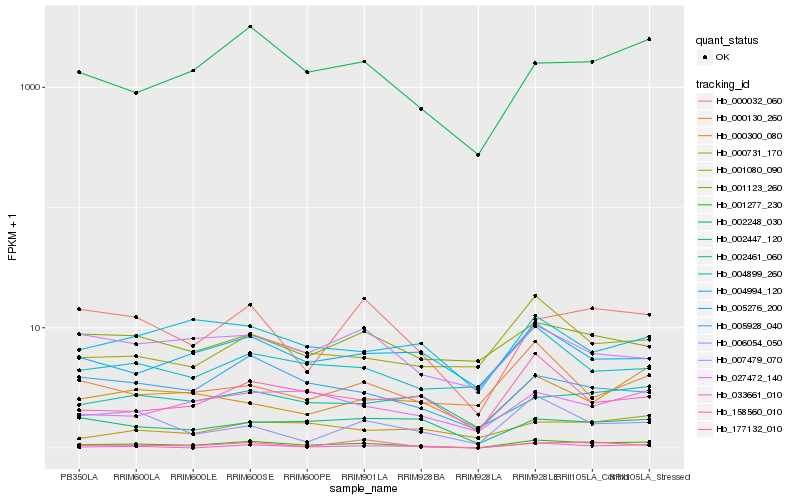
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_230 |
0.0 |
- |
- |
- |
| 2 |
Hb_002248_030 |
0.1626112717 |
- |
- |
metallothionein-like protein [Hevea brasiliensis] |
| 3 |
Hb_000032_060 |
0.1670681654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005928_040 |
0.1797308699 |
- |
- |
phosphoinositide phosphatase family protein [Populus trichocarpa] |
| 5 |
Hb_027472_140 |
0.1940466007 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646176 [Jatropha curcas] |
| 6 |
Hb_177132_010 |
0.1941219621 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_158560_010 |
0.1975168685 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 8 |
Hb_004899_260 |
0.1980408225 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 9 |
Hb_001080_090 |
0.2031822213 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002447_120 |
0.2033127953 |
- |
- |
hypothetical protein O988_01801, partial [Pseudogymnoascus pannorum VKM F-3808] |
| 11 |
Hb_002461_060 |
0.2045206658 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 12 |
Hb_007479_070 |
0.2047201803 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 13 |
Hb_000300_080 |
0.2055728246 |
- |
- |
PREDICTED: probable S-sulfocysteine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_005276_200 |
0.2061728462 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_033661_010 |
0.2079768775 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 16 |
Hb_004994_120 |
0.2099969537 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 13-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000130_260 |
0.2123226244 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006054_050 |
0.212398456 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 19 |
Hb_001123_260 |
0.2126951111 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 20 |
Hb_000731_170 |
0.2136633276 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |