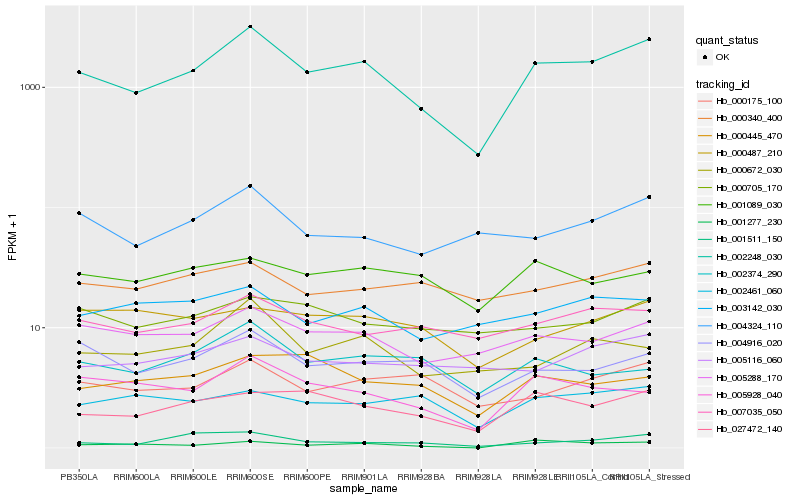
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002248_030 |
0.0 |
- |
- |
metallothionein-like protein [Hevea brasiliensis] |
| 2 |
Hb_027472_140 |
0.1267502937 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646176 [Jatropha curcas] |
| 3 |
Hb_005288_170 |
0.1339639074 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08610 [Populus euphratica] |
| 4 |
Hb_005928_040 |
0.1400783723 |
- |
- |
phosphoinositide phosphatase family protein [Populus trichocarpa] |
| 5 |
Hb_004324_110 |
0.143781885 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 6 |
Hb_000672_030 |
0.1450603188 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004916_020 |
0.1460869084 |
- |
- |
PREDICTED: DNA ligase 4 [Jatropha curcas] |
| 8 |
Hb_000175_100 |
0.149832495 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 9 |
Hb_007035_050 |
0.1508545504 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 10 |
Hb_005116_060 |
0.155997051 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003142_030 |
0.1562193824 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002461_060 |
0.1573327066 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 13 |
Hb_000445_470 |
0.1596507892 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 14 |
Hb_001511_150 |
0.159768073 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 15 |
Hb_000705_170 |
0.1598131538 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 16 |
Hb_001089_030 |
0.1606627977 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 17 |
Hb_000487_210 |
0.1610606488 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 18 |
Hb_000340_400 |
0.1617567303 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 19 |
Hb_002374_290 |
0.1623381176 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001277_230 |
0.1626112717 |
- |
- |
- |