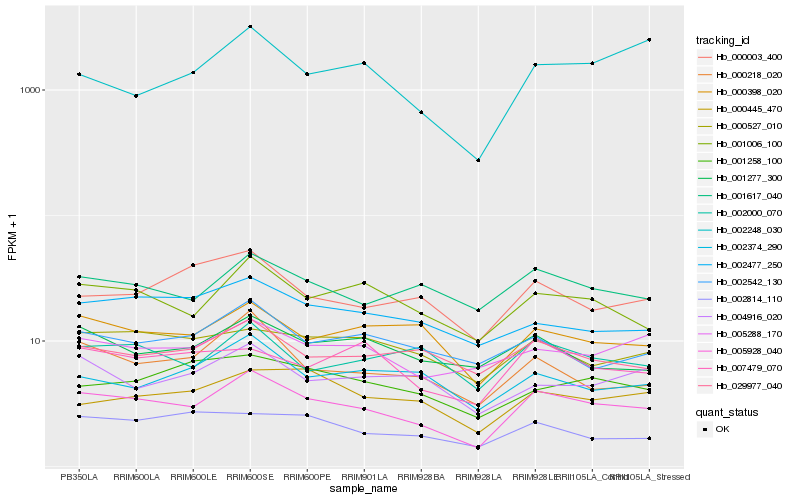
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005928_040 |
0.0 |
- |
- |
phosphoinositide phosphatase family protein [Populus trichocarpa] |
| 2 |
Hb_001006_100 |
0.1107136797 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_000218_020 |
0.1220299488 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 4 |
Hb_000398_020 |
0.1228919322 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001617_040 |
0.1261989506 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_002000_070 |
0.1294248464 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 5-formyltetrahydrofolate cyclo-ligase-like protein COG0212 [Jatropha curcas] |
| 7 |
Hb_005288_170 |
0.1302687391 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08610 [Populus euphratica] |
| 8 |
Hb_029977_040 |
0.1325338645 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 9 |
Hb_000527_010 |
0.1327690365 |
- |
- |
katanin P80 subunit, putative [Ricinus communis] |
| 10 |
Hb_000445_470 |
0.1330524347 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 11 |
Hb_000003_400 |
0.1339333125 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_001277_300 |
0.1351363091 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 13 |
Hb_002542_130 |
0.1355443893 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 14 |
Hb_002477_250 |
0.1359216655 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 15 |
Hb_007479_070 |
0.1367979479 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 16 |
Hb_001258_100 |
0.1383285263 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_002374_290 |
0.1389400467 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004916_020 |
0.1390437616 |
- |
- |
PREDICTED: DNA ligase 4 [Jatropha curcas] |
| 19 |
Hb_002248_030 |
0.1400783723 |
- |
- |
metallothionein-like protein [Hevea brasiliensis] |
| 20 |
Hb_002814_110 |
0.141205454 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |