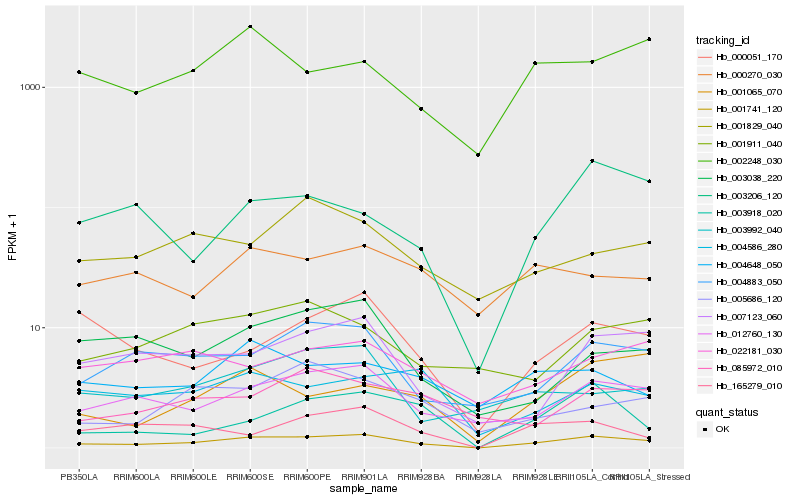
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001741_120 |
0.0 |
- |
- |
hypothetical protein CARUB_v10011543mg, partial [Capsella rubella] |
| 2 |
Hb_012760_130 |
0.1598925183 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 3 |
Hb_004883_050 |
0.1861580917 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 4 |
Hb_007123_060 |
0.192622 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 5 |
Hb_000051_170 |
0.2000004486 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 6 |
Hb_003206_120 |
0.2093136678 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 7 |
Hb_003992_040 |
0.2094403743 |
- |
- |
Post-illumination chlorophyll fluorescence increase isoform 5 [Theobroma cacao] |
| 8 |
Hb_002248_030 |
0.2100138577 |
- |
- |
metallothionein-like protein [Hevea brasiliensis] |
| 9 |
Hb_003918_020 |
0.2103882612 |
- |
- |
- |
| 10 |
Hb_001065_070 |
0.2118190924 |
- |
- |
- |
| 11 |
Hb_003038_220 |
0.2121634944 |
- |
- |
eukaryotic translation initiation factor 2 gamma subunit, putative [Ricinus communis] |
| 12 |
Hb_004648_050 |
0.2125413371 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_165279_010 |
0.2143962249 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 14 |
Hb_001911_040 |
0.2145548457 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 15 |
Hb_000270_030 |
0.2147809483 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0015s07640g [Populus trichocarpa] |
| 16 |
Hb_005686_120 |
0.2150480602 |
- |
- |
- |
| 17 |
Hb_001829_040 |
0.2160447032 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 18 |
Hb_004586_280 |
0.2165265514 |
- |
- |
PREDICTED: GEM-like protein 1 [Jatropha curcas] |
| 19 |
Hb_085972_010 |
0.2174214212 |
transcription factor |
TF Family: NAC |
NAC transcription factor 066 [Jatropha curcas] |
| 20 |
Hb_022181_030 |
0.2178166409 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |