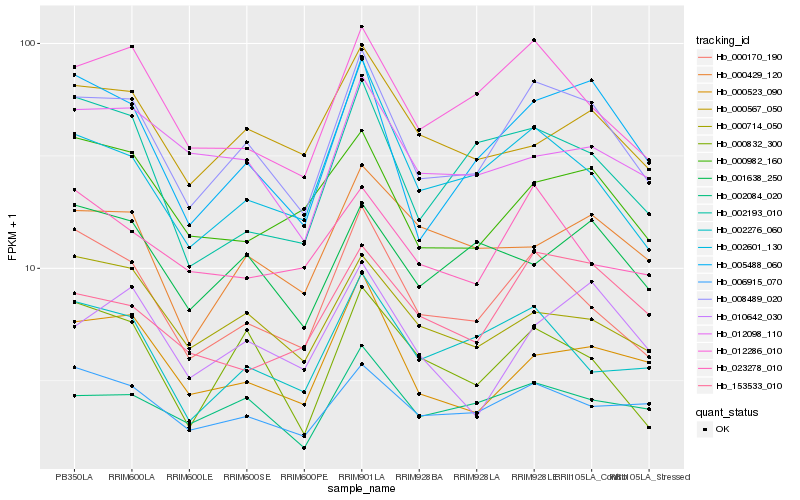
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008489_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644663 [Jatropha curcas] |
| 2 |
Hb_005488_060 |
0.0913507226 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_000170_190 |
0.0980917262 |
- |
- |
PREDICTED: uncharacterized protein LOC105638988 [Jatropha curcas] |
| 4 |
Hb_012286_010 |
0.100431936 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit C-like [Jatropha curcas] |
| 5 |
Hb_002084_020 |
0.1058521667 |
- |
- |
hypothetical protein JCGZ_04066 [Jatropha curcas] |
| 6 |
Hb_002601_130 |
0.1064585736 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 7 |
Hb_006915_070 |
0.1119018751 |
- |
- |
hypothetical protein CAPTEDRAFT_111329, partial [Capitella teleta] |
| 8 |
Hb_000523_090 |
0.1153630089 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 9 |
Hb_000982_160 |
0.1186468085 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 10 |
Hb_002276_060 |
0.1192047257 |
- |
- |
hypothetical protein CISIN_1g002019mg [Citrus sinensis] |
| 11 |
Hb_000714_050 |
0.1217403651 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 12 |
Hb_002193_010 |
0.121813434 |
- |
- |
PREDICTED: SUN domain-containing protein 3-like [Jatropha curcas] |
| 13 |
Hb_153533_010 |
0.1229608262 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X3 [Jatropha curcas] |
| 14 |
Hb_000567_050 |
0.1234761531 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 15 |
Hb_000832_300 |
0.1238885124 |
- |
- |
- |
| 16 |
Hb_010642_030 |
0.1269463181 |
- |
- |
zinc phosphodiesterase, putative [Ricinus communis] |
| 17 |
Hb_023278_010 |
0.1273773124 |
- |
- |
Chaperone protein DnaJ [Glycine soja] |
| 18 |
Hb_012098_110 |
0.1275065762 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 19 |
Hb_001638_250 |
0.1281734545 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05600 [Jatropha curcas] |
| 20 |
Hb_000429_120 |
0.1283909789 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |