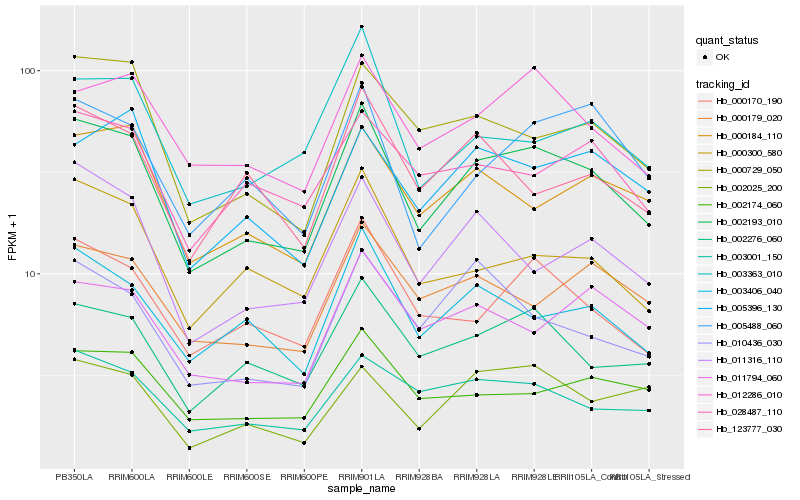
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002193_010 |
0.0 |
- |
- |
PREDICTED: SUN domain-containing protein 3-like [Jatropha curcas] |
| 2 |
Hb_002276_060 |
0.0934262171 |
- |
- |
hypothetical protein CISIN_1g002019mg [Citrus sinensis] |
| 3 |
Hb_012286_010 |
0.0994461726 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit C-like [Jatropha curcas] |
| 4 |
Hb_000170_190 |
0.1005802793 |
- |
- |
PREDICTED: uncharacterized protein LOC105638988 [Jatropha curcas] |
| 5 |
Hb_003406_040 |
0.103439658 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 6 |
Hb_003001_150 |
0.1038544682 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000729_050 |
0.107239264 |
- |
- |
Pinin, putative [Ricinus communis] |
| 8 |
Hb_000184_110 |
0.1082596514 |
- |
- |
hypothetical protein F383_18769 [Gossypium arboreum] |
| 9 |
Hb_011316_110 |
0.1084684454 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 10 |
Hb_000300_580 |
0.1098977613 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 11 |
Hb_002174_060 |
0.1126097471 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 12 |
Hb_005396_130 |
0.1127327406 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 13 |
Hb_000179_020 |
0.1128483754 |
- |
- |
amidase, putative [Ricinus communis] |
| 14 |
Hb_005488_060 |
0.1142449752 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_003363_010 |
0.1164295016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002025_200 |
0.1167684317 |
- |
- |
- |
| 17 |
Hb_011794_060 |
0.1172019914 |
- |
- |
PREDICTED: beta-amylase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_028487_110 |
0.1178311501 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 19 |
Hb_010436_030 |
0.119823947 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 20 |
Hb_123777_030 |
0.12053264 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA helicase INO80 isoform X2 [Jatropha curcas] |