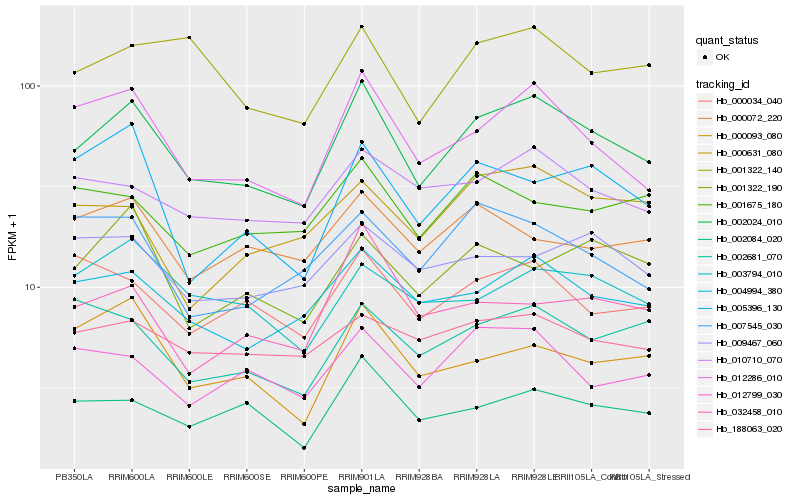
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002024_010 |
0.0 |
- |
- |
hypothetical protein B456_003G0564002, partial [Gossypium raimondii] |
| 2 |
Hb_012286_010 |
0.0850732516 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit C-like [Jatropha curcas] |
| 3 |
Hb_004994_380 |
0.0924538529 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 4 |
Hb_002084_020 |
0.1095383284 |
- |
- |
hypothetical protein JCGZ_04066 [Jatropha curcas] |
| 5 |
Hb_010710_070 |
0.1107025604 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_012799_030 |
0.116065941 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g55630-like [Jatropha curcas] |
| 7 |
Hb_001322_140 |
0.1162935322 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_188063_020 |
0.1171469119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001322_190 |
0.12037013 |
- |
- |
PREDICTED: uncharacterized protein LOC105632381 [Jatropha curcas] |
| 10 |
Hb_000034_040 |
0.1204819716 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 11 |
Hb_003794_010 |
0.1206809401 |
- |
- |
PREDICTED: uncharacterized protein LOC105641013 [Jatropha curcas] |
| 12 |
Hb_002681_070 |
0.1238373682 |
- |
- |
PREDICTED: uncharacterized protein LOC105633833 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007545_030 |
0.1238922448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001675_180 |
0.1240751381 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 15 |
Hb_000631_080 |
0.1245256007 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Jatropha curcas] |
| 16 |
Hb_000072_220 |
0.124693395 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02490, mitochondrial [Jatropha curcas] |
| 17 |
Hb_005396_130 |
0.1247324347 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 18 |
Hb_000093_080 |
0.1247472363 |
- |
- |
hypothetical protein POPTR_0008s09730g [Populus trichocarpa] |
| 19 |
Hb_009467_060 |
0.1247976492 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_032458_010 |
0.1263288404 |
- |
- |
hypothetical protein JCGZ_21107 [Jatropha curcas] |