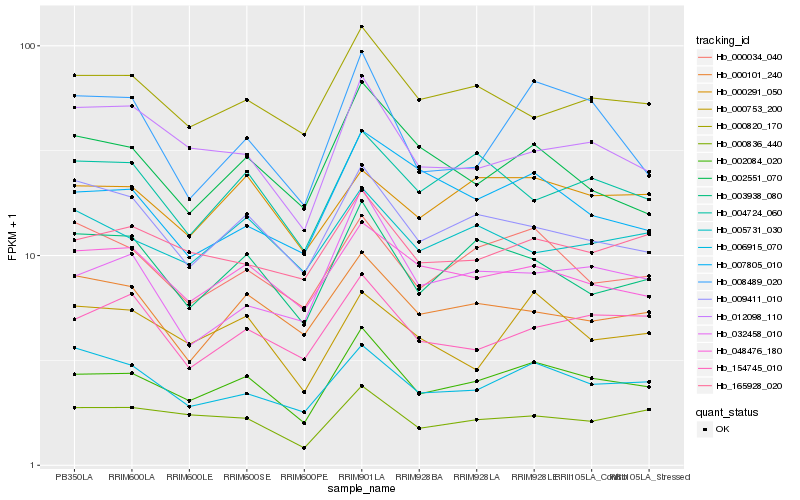
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002084_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_04066 [Jatropha curcas] |
| 2 |
Hb_000836_440 |
0.0888505451 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_048476_180 |
0.0896800602 |
- |
- |
PREDICTED: nuclear pore complex protein NUP155 [Jatropha curcas] |
| 4 |
Hb_003938_080 |
0.0909435285 |
- |
- |
PREDICTED: uncharacterized protein LOC105644554 [Jatropha curcas] |
| 5 |
Hb_165928_020 |
0.0935469889 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000820_170 |
0.0942294879 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 7 |
Hb_000034_040 |
0.0947214203 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 8 |
Hb_006915_070 |
0.0960572441 |
- |
- |
hypothetical protein CAPTEDRAFT_111329, partial [Capitella teleta] |
| 9 |
Hb_009411_010 |
0.0981285684 |
- |
- |
PREDICTED: uncharacterized protein LOC105645272 [Jatropha curcas] |
| 10 |
Hb_154745_010 |
0.098915254 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 11 |
Hb_004724_060 |
0.1000986955 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000291_050 |
0.1004194436 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012098_110 |
0.1006365929 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 14 |
Hb_032458_010 |
0.1015203833 |
- |
- |
hypothetical protein JCGZ_21107 [Jatropha curcas] |
| 15 |
Hb_000101_240 |
0.1022554458 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 16 |
Hb_002551_070 |
0.1028985019 |
- |
- |
hypothetical protein POPTR_0019s10130g [Populus trichocarpa] |
| 17 |
Hb_007805_010 |
0.1040200402 |
- |
- |
PREDICTED: ribosome biogenesis protein BMS1 homolog [Jatropha curcas] |
| 18 |
Hb_000753_200 |
0.1047977697 |
- |
- |
PREDICTED: putative ATP-dependent helicase hrq1 [Jatropha curcas] |
| 19 |
Hb_005731_030 |
0.1058261875 |
- |
- |
PREDICTED: UPF0400 protein C337.03 [Jatropha curcas] |
| 20 |
Hb_008489_020 |
0.1058521667 |
- |
- |
PREDICTED: uncharacterized protein LOC105644663 [Jatropha curcas] |