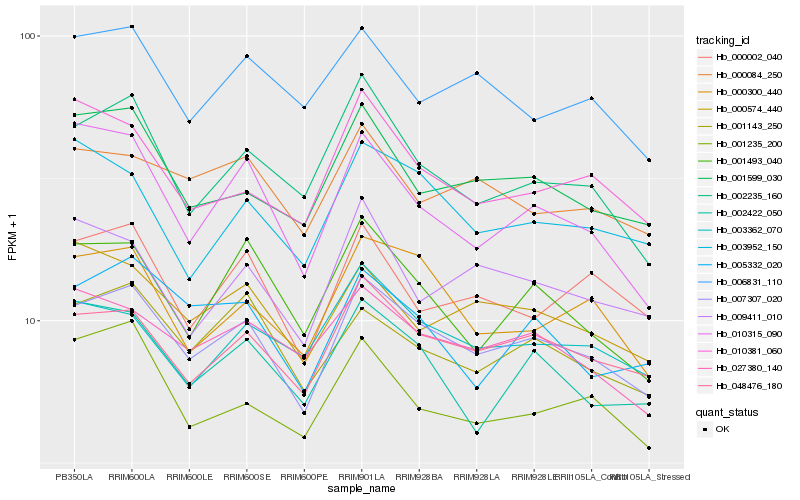
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007307_020 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_048476_180 |
0.0503401227 |
- |
- |
PREDICTED: nuclear pore complex protein NUP155 [Jatropha curcas] |
| 3 |
Hb_001143_250 |
0.0691969966 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 4 |
Hb_001599_030 |
0.0729525473 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 5 |
Hb_000300_440 |
0.073053129 |
- |
- |
MIF4G domain and MA3 domain-containing protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_027380_140 |
0.0734321022 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002235_160 |
0.0738722753 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 8 |
Hb_000574_440 |
0.0753071089 |
- |
- |
PREDICTED: uncharacterized protein LOC105634833 [Jatropha curcas] |
| 9 |
Hb_000084_250 |
0.0767651705 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 10 |
Hb_003362_070 |
0.078579473 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 11 |
Hb_000002_040 |
0.0787225541 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 12 |
Hb_005332_020 |
0.0787490733 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_003952_150 |
0.0804895817 |
- |
- |
PREDICTED: cactin [Jatropha curcas] |
| 14 |
Hb_002422_050 |
0.0807341086 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 15 |
Hb_010315_090 |
0.0807819286 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 16 |
Hb_010381_060 |
0.0816858811 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 17 |
Hb_001493_040 |
0.0820875663 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 18 |
Hb_009411_010 |
0.082152545 |
- |
- |
PREDICTED: uncharacterized protein LOC105645272 [Jatropha curcas] |
| 19 |
Hb_001235_200 |
0.0825908364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006831_110 |
0.0827686745 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |