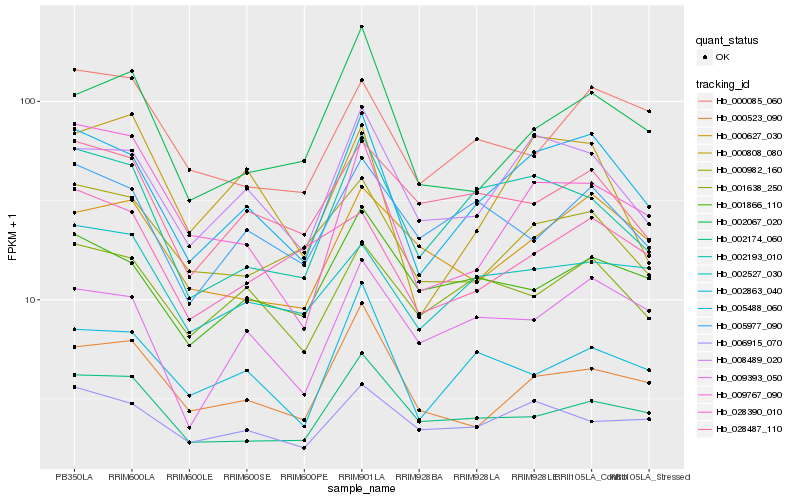
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005488_060 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_008489_020 |
0.0913507226 |
- |
- |
PREDICTED: uncharacterized protein LOC105644663 [Jatropha curcas] |
| 3 |
Hb_002193_010 |
0.1142449752 |
- |
- |
PREDICTED: SUN domain-containing protein 3-like [Jatropha curcas] |
| 4 |
Hb_009393_050 |
0.1200940058 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 5 |
Hb_001638_250 |
0.1229133014 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05600 [Jatropha curcas] |
| 6 |
Hb_000982_160 |
0.124097956 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 7 |
Hb_002863_040 |
0.1248331077 |
- |
- |
PREDICTED: uncharacterized protein LOC105630268 [Jatropha curcas] |
| 8 |
Hb_002527_030 |
0.1285232354 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1-like [Jatropha curcas] |
| 9 |
Hb_000808_080 |
0.1299884384 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 3 [Jatropha curcas] |
| 10 |
Hb_000523_090 |
0.132674658 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 11 |
Hb_005977_090 |
0.1339983463 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g16250 [Jatropha curcas] |
| 12 |
Hb_001866_110 |
0.1348213979 |
- |
- |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 13 |
Hb_009767_090 |
0.1361337608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002174_060 |
0.1367696488 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 15 |
Hb_028487_110 |
0.136869563 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_006915_070 |
0.1379625856 |
- |
- |
hypothetical protein CAPTEDRAFT_111329, partial [Capitella teleta] |
| 17 |
Hb_002067_020 |
0.1388994398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000085_060 |
0.1391075949 |
- |
- |
PREDICTED: uncharacterized protein LOC105633020 [Jatropha curcas] |
| 19 |
Hb_028390_010 |
0.1392238869 |
- |
- |
PREDICTED: filament-like plant protein 3 [Populus euphratica] |
| 20 |
Hb_000627_030 |
0.1392904977 |
- |
- |
amine oxidase, putative [Ricinus communis] |