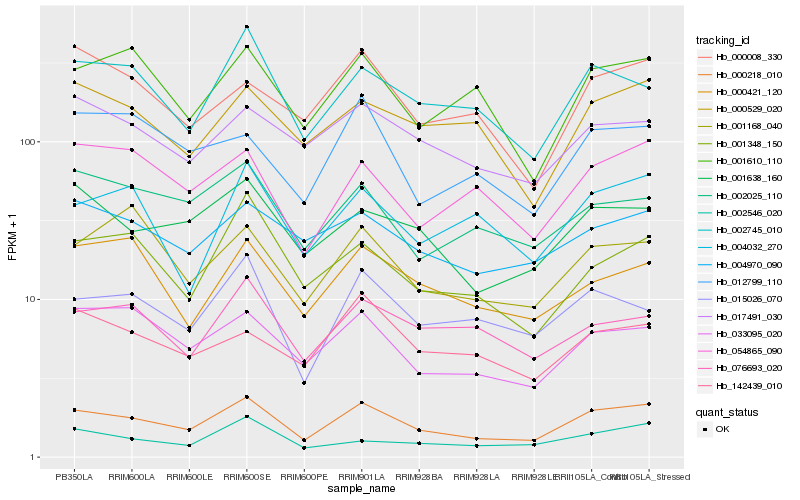
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000218_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_033095_020 |
0.1093126824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001638_160 |
0.1096240141 |
- |
- |
PREDICTED: uncharacterized protein LOC105642785 [Jatropha curcas] |
| 4 |
Hb_002745_010 |
0.1115464547 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 5 |
Hb_004970_090 |
0.1152206176 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 6 |
Hb_017491_030 |
0.116113689 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 7 |
Hb_054865_090 |
0.1163708933 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 8 |
Hb_000529_020 |
0.1179944848 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 9 |
Hb_015026_070 |
0.1185258079 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001610_110 |
0.119125309 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 11 |
Hb_002025_110 |
0.1207737905 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_001348_150 |
0.1232404682 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 13 |
Hb_076693_020 |
0.1247659885 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 14 |
Hb_002546_020 |
0.1252664793 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Malus domestica] |
| 15 |
Hb_000421_120 |
0.1258179773 |
- |
- |
PREDICTED: NEDD8-specific protease 1 [Jatropha curcas] |
| 16 |
Hb_142439_010 |
0.1258353978 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 17 |
Hb_000008_330 |
0.1262616174 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 18 |
Hb_004032_270 |
0.1268839167 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_001168_040 |
0.1269673033 |
- |
- |
- |
| 20 |
Hb_012799_110 |
0.128770642 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |