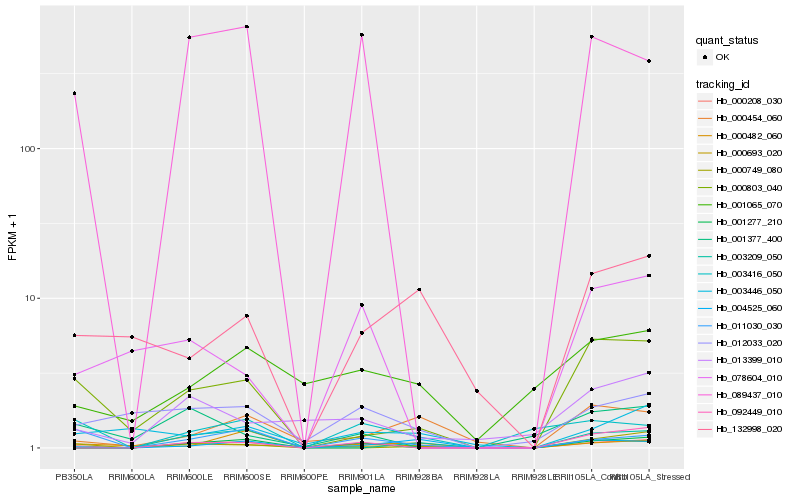
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000208_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_089437_010 |
0.2237155905 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 10-like isoform X3 [Jatropha curcas] |
| 3 |
Hb_011030_030 |
0.2527424014 |
- |
- |
PLC-like phosphodiesterases superfamily protein [Theobroma cacao] |
| 4 |
Hb_001277_210 |
0.2582583083 |
- |
- |
- |
| 5 |
Hb_012033_020 |
0.2788885556 |
- |
- |
- |
| 6 |
Hb_000803_040 |
0.2870301846 |
- |
- |
- |
| 7 |
Hb_004525_060 |
0.2993455543 |
- |
- |
- |
| 8 |
Hb_003416_050 |
0.3059817413 |
- |
- |
- |
| 9 |
Hb_000693_020 |
0.308875565 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 10 |
Hb_013399_010 |
0.3138017562 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 11 |
Hb_092449_010 |
0.3139454106 |
- |
- |
- |
| 12 |
Hb_000454_060 |
0.3175500099 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |
| 13 |
Hb_003446_050 |
0.3201895039 |
- |
- |
hypothetical protein B456_002G034000, partial [Gossypium raimondii] |
| 14 |
Hb_001065_070 |
0.3212104329 |
- |
- |
- |
| 15 |
Hb_132998_020 |
0.3232363388 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000749_080 |
0.3236209471 |
- |
- |
Alpha-expansin 12 precursor, putative [Ricinus communis] |
| 17 |
Hb_003209_050 |
0.3268608227 |
- |
- |
hypothetical protein JCGZ_15683 [Jatropha curcas] |
| 18 |
Hb_000482_060 |
0.3303354412 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_07664 [Jatropha curcas] |
| 19 |
Hb_001377_400 |
0.3332661899 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 20 |
Hb_078604_010 |
0.3361351172 |
- |
- |
- |