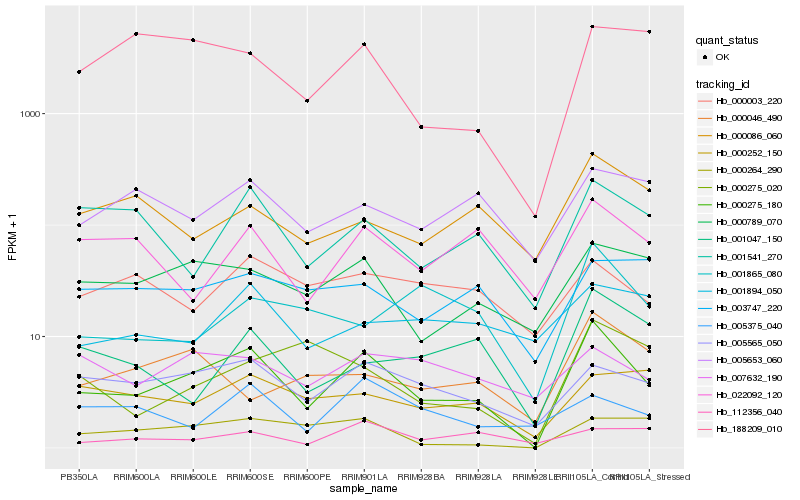
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000275_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_005565_050 |
0.2271447682 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 3 |
Hb_000789_070 |
0.2318222025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001541_270 |
0.241651796 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 5 |
Hb_022092_120 |
0.2526626328 |
- |
- |
PREDICTED: uncharacterized protein LOC105646971 [Jatropha curcas] |
| 6 |
Hb_000264_290 |
0.2557154686 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 7 |
Hb_188209_010 |
0.2592380678 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001047_150 |
0.2600056251 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 9 |
Hb_001865_080 |
0.2610569457 |
- |
- |
enoyl-CoA hydratase/isomerase family protein [Populus trichocarpa] |
| 10 |
Hb_000086_060 |
0.2634611153 |
- |
- |
PREDICTED: TOM1-like protein 2 isoform X2 [Populus euphratica] |
| 11 |
Hb_005653_060 |
0.2639341537 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 12 |
Hb_000252_150 |
0.2704870601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007632_190 |
0.2706451361 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic isoform X5 [Camelina sativa] |
| 14 |
Hb_003747_220 |
0.2715692679 |
- |
- |
PREDICTED: syntaxin-22-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001894_050 |
0.2718512161 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 2 [Jatropha curcas] |
| 16 |
Hb_000003_220 |
0.272386989 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 17 |
Hb_000046_490 |
0.2727518353 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 18 |
Hb_005375_040 |
0.2735807963 |
- |
- |
dicer-1, putative [Ricinus communis] |
| 19 |
Hb_000275_020 |
0.2736329583 |
- |
- |
PREDICTED: uncharacterized protein LOC105632656 [Jatropha curcas] |
| 20 |
Hb_112356_040 |
0.2750145197 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |