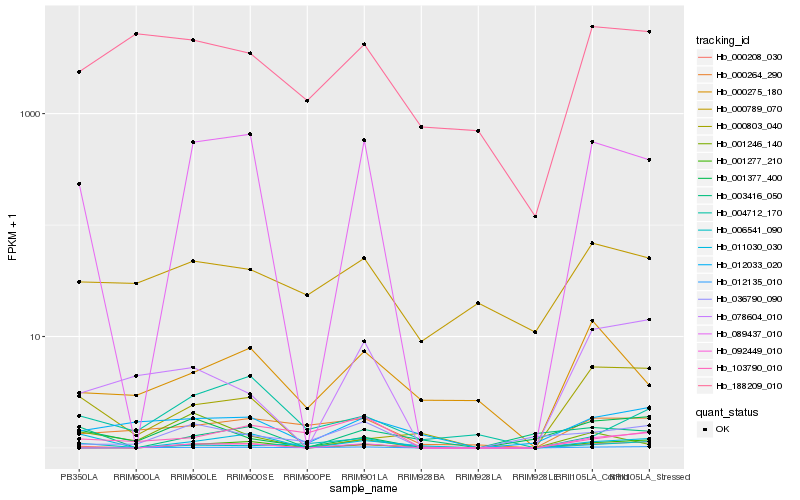
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089437_010 |
0.0 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 10-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_000208_030 |
0.2237155905 |
- |
- |
- |
| 3 |
Hb_001277_210 |
0.2716196747 |
- |
- |
- |
| 4 |
Hb_011030_030 |
0.2739988071 |
- |
- |
PLC-like phosphodiesterases superfamily protein [Theobroma cacao] |
| 5 |
Hb_003416_050 |
0.3343687293 |
- |
- |
- |
| 6 |
Hb_036790_090 |
0.3409008796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000275_180 |
0.3428257129 |
- |
- |
- |
| 8 |
Hb_012033_020 |
0.3467036055 |
- |
- |
- |
| 9 |
Hb_000264_290 |
0.3529007836 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 10 |
Hb_078604_010 |
0.3531729204 |
- |
- |
- |
| 11 |
Hb_001377_400 |
0.3562845692 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 12 |
Hb_103790_010 |
0.3615156099 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 13 |
Hb_001246_140 |
0.3627374041 |
- |
- |
- |
| 14 |
Hb_012135_010 |
0.3679056444 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 15 |
Hb_004712_170 |
0.3746856134 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 16 |
Hb_092449_010 |
0.3757565045 |
- |
- |
- |
| 17 |
Hb_000789_070 |
0.3781988996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006541_090 |
0.3800702817 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 19 |
Hb_000803_040 |
0.3804136585 |
- |
- |
- |
| 20 |
Hb_188209_010 |
0.3812910363 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |