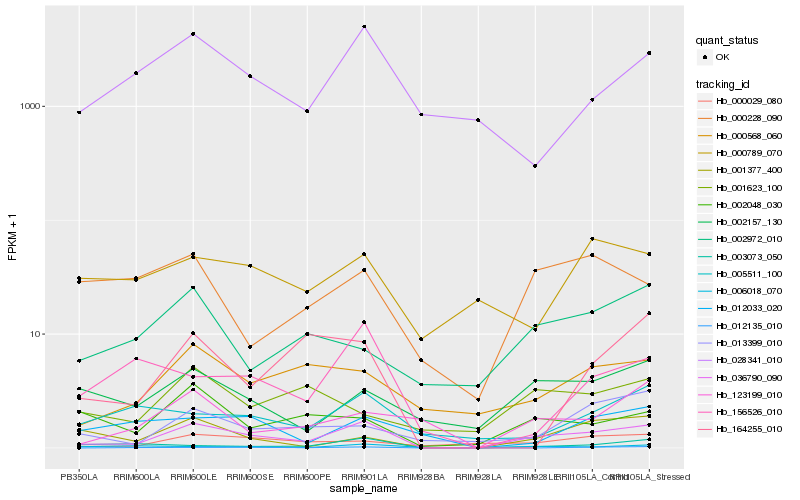
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036790_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000568_060 |
0.2676284429 |
- |
- |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 3 |
Hb_000029_080 |
0.2713275491 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 4 |
Hb_002157_130 |
0.2771366869 |
- |
- |
- |
| 5 |
Hb_002048_030 |
0.2865541527 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 6 |
Hb_123199_010 |
0.2878208285 |
- |
- |
SRK protein [Brassica cretica] |
| 7 |
Hb_012135_010 |
0.2891650544 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 8 |
Hb_013399_010 |
0.2900544135 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 9 |
Hb_028341_010 |
0.2917787763 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 10 |
Hb_001377_400 |
0.2927903104 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 11 |
Hb_003073_050 |
0.293794244 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 12 |
Hb_005511_100 |
0.2974361953 |
- |
- |
PREDICTED: uncharacterized protein LOC105645493 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006018_070 |
0.2984866836 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 14 |
Hb_002972_010 |
0.298581787 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 15 |
Hb_156526_010 |
0.3036074047 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 16 |
Hb_012033_020 |
0.3041178952 |
- |
- |
- |
| 17 |
Hb_001623_100 |
0.3047346326 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 18 |
Hb_000228_090 |
0.3051699049 |
- |
- |
PREDICTED: protein PAM68, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000789_070 |
0.3055102779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_164255_010 |
0.3057316605 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |