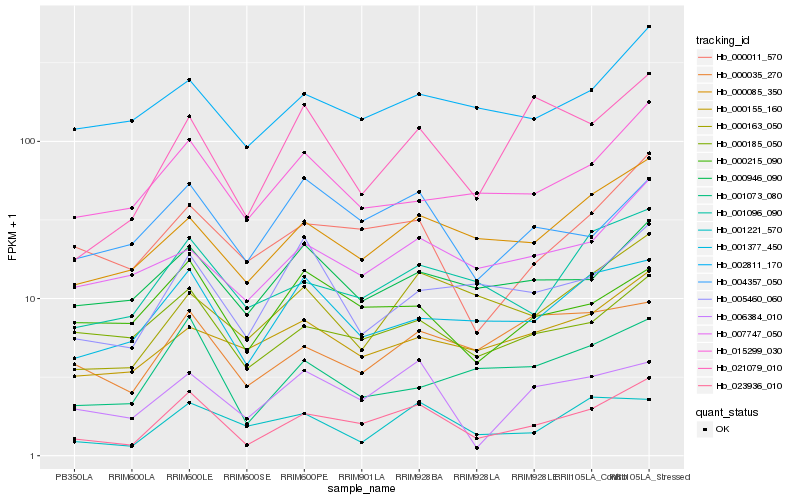
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023936_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000085_350 |
0.1501941002 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000011_570 |
0.1510679749 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 4 |
Hb_001096_090 |
0.1514477982 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001377_450 |
0.1584264777 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000185_050 |
0.1643787469 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 7 |
Hb_001073_080 |
0.1646144608 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_015299_030 |
0.1648343226 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 9 |
Hb_002811_170 |
0.1677310886 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 10 |
Hb_000163_050 |
0.1677953936 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 11 |
Hb_007747_050 |
0.1697277321 |
- |
- |
Peptidyl-prolyl cis-trans isomerase cypE, putative [Ricinus communis] |
| 12 |
Hb_006384_010 |
0.1708580496 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_000946_090 |
0.1724484951 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000035_270 |
0.1746095176 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |
| 15 |
Hb_021079_010 |
0.1759178694 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 16 |
Hb_004357_050 |
0.1761388608 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 17 |
Hb_001221_570 |
0.1764798706 |
- |
- |
- |
| 18 |
Hb_000155_160 |
0.1767892958 |
- |
- |
hypothetical protein POPTR_0011s05880g [Populus trichocarpa] |
| 19 |
Hb_005460_060 |
0.1769016917 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 20 |
Hb_000215_090 |
0.1775718686 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |