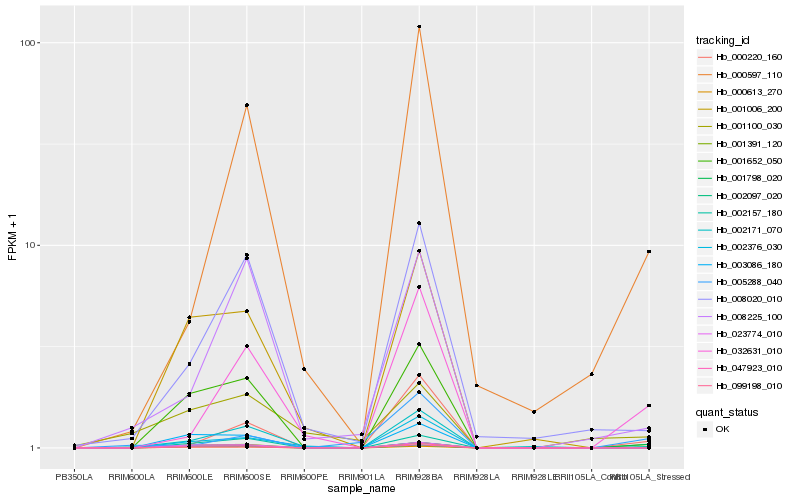
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002097_020 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 2 |
Hb_001391_120 |
0.2320335999 |
- |
- |
Acetyl esterase, putative [Ricinus communis] |
| 3 |
Hb_001100_030 |
0.296970856 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 4 |
Hb_002376_030 |
0.3043717677 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_001652_050 |
0.3065398684 |
- |
- |
- |
| 6 |
Hb_005288_040 |
0.3067605519 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_032631_010 |
0.3074958164 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 8 |
Hb_003086_180 |
0.3143173608 |
- |
- |
PREDICTED: RING-H2 finger protein ATL70-like [Pyrus x bretschneideri] |
| 9 |
Hb_023774_010 |
0.3146911568 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 10 |
Hb_008020_010 |
0.3149129831 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like [Jatropha curcas] |
| 11 |
Hb_000613_270 |
0.3158937472 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 12 |
Hb_000220_160 |
0.3171308159 |
- |
- |
hypothetical protein B456_003G184800 [Gossypium raimondii] |
| 13 |
Hb_008225_100 |
0.3181094673 |
- |
- |
- |
| 14 |
Hb_001798_020 |
0.3185686858 |
- |
- |
PREDICTED: uncharacterized protein LOC104594032 [Nelumbo nucifera] |
| 15 |
Hb_002157_180 |
0.318909356 |
- |
- |
separase, putative [Ricinus communis] |
| 16 |
Hb_099198_010 |
0.3189275079 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At2g25790 isoform X2 [Nicotiana tomentosiformis] |
| 17 |
Hb_002171_070 |
0.3192537813 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |
| 18 |
Hb_000597_110 |
0.3194613926 |
- |
- |
PREDICTED: VQ motif-containing protein 22-like [Jatropha curcas] |
| 19 |
Hb_047923_010 |
0.3217776078 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 20 |
Hb_001006_200 |
0.3222140924 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |