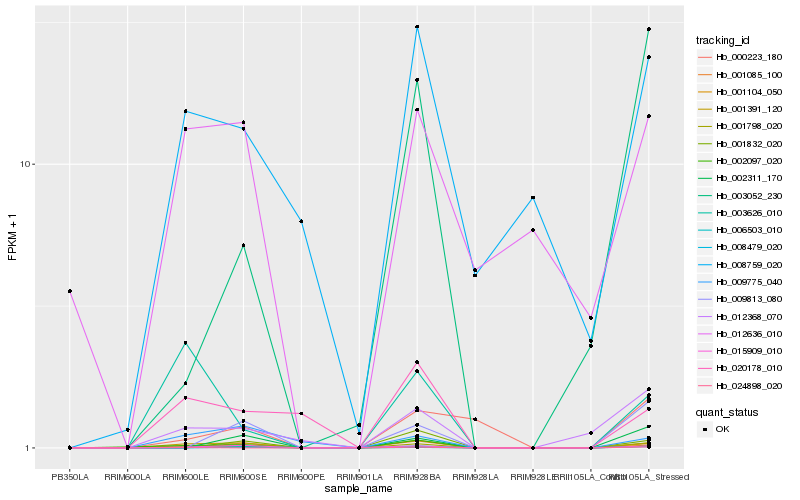
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001798_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104594032 [Nelumbo nucifera] |
| 2 |
Hb_001104_050 |
0.1272371748 |
- |
- |
- |
| 3 |
Hb_001391_120 |
0.1391207134 |
- |
- |
Acetyl esterase, putative [Ricinus communis] |
| 4 |
Hb_012368_070 |
0.2670345077 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_008479_020 |
0.2970584718 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 6 |
Hb_003052_230 |
0.3005815296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_024898_020 |
0.3036181053 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_015909_010 |
0.3041341838 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_009813_080 |
0.309762419 |
- |
- |
hypothetical protein CISIN_1g045991mg [Citrus sinensis] |
| 10 |
Hb_002311_170 |
0.3145919964 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Eucalyptus grandis] |
| 11 |
Hb_008759_020 |
0.3160844934 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 12 |
Hb_002097_020 |
0.3185686858 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 13 |
Hb_001085_100 |
0.3234069924 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_003626_010 |
0.3235312832 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 15 |
Hb_020178_010 |
0.3290367989 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 16 |
Hb_006503_010 |
0.3316902564 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_009775_040 |
0.341216288 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Vitis vinifera] |
| 18 |
Hb_012636_010 |
0.3431189404 |
- |
- |
- |
| 19 |
Hb_000223_180 |
0.3434070531 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 20 |
Hb_001832_020 |
0.3464243553 |
- |
- |
PREDICTED: uncharacterized protein LOC104584029 [Brachypodium distachyon] |