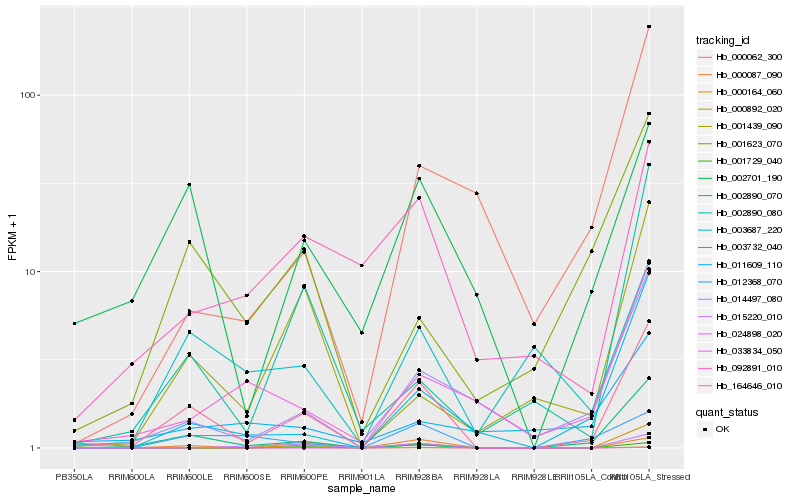
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164646_010 |
0.0 |
- |
- |
PREDICTED: acidic endochitinase-like [Pyrus x bretschneideri] |
| 2 |
Hb_014497_080 |
0.2610431467 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 3 |
Hb_002701_190 |
0.2610898999 |
- |
- |
PREDICTED: uncharacterized protein LOC105646742 [Jatropha curcas] |
| 4 |
Hb_000087_090 |
0.2651145515 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 5 |
Hb_002890_070 |
0.2765211775 |
- |
- |
ATP-dependent RNA helicase family protein [Populus trichocarpa] |
| 6 |
Hb_003732_040 |
0.2796253089 |
- |
- |
putative esterase family protein [Populus trichocarpa] |
| 7 |
Hb_000164_060 |
0.2887983282 |
- |
- |
- |
| 8 |
Hb_001623_070 |
0.2913923257 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 9 |
Hb_012368_070 |
0.292107787 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_001729_040 |
0.2935081741 |
- |
- |
hypothetical protein glysoja_049826, partial [Glycine soja] |
| 11 |
Hb_002890_080 |
0.2975834307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_015220_010 |
0.3010498248 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 13 |
Hb_001439_090 |
0.3026006667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000892_020 |
0.3061602678 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_024898_020 |
0.3076726697 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_003687_220 |
0.3083608869 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 17 |
Hb_033834_050 |
0.3098456335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011609_110 |
0.3102920331 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 19 |
Hb_092891_010 |
0.312429778 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 20 |
Hb_000062_300 |
0.3166000256 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |