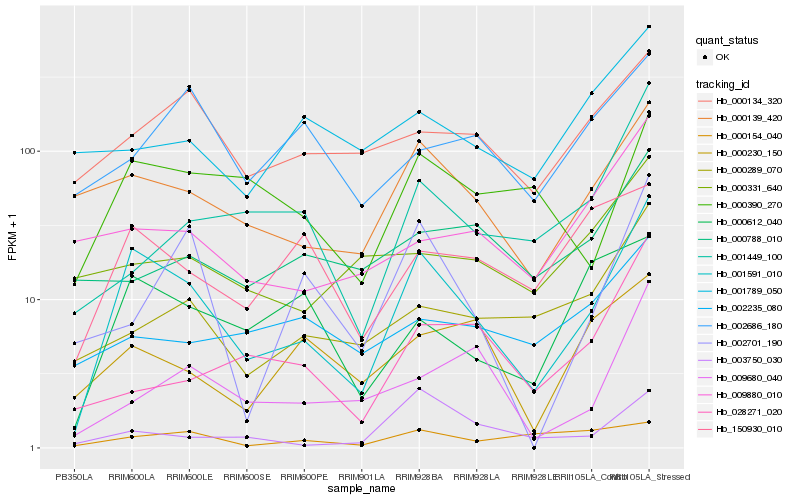
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001591_010 |
0.0 |
transcription factor |
TF Family: MYB |
myb domain protein [Hevea brasiliensis] |
| 2 |
Hb_000139_420 |
0.2199231318 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 3 |
Hb_000390_270 |
0.2541631269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000612_040 |
0.2600240831 |
- |
- |
hypothetical protein OsJ_16945 [Oryza sativa Japonica Group] |
| 5 |
Hb_000289_070 |
0.2671991151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003750_030 |
0.2690891077 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 7 |
Hb_000134_320 |
0.2712688609 |
- |
- |
hypothetical protein PRUPE_ppa013450mg [Prunus persica] |
| 8 |
Hb_150930_010 |
0.2717603744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009880_010 |
0.2745322809 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 10 |
Hb_002701_190 |
0.2745360609 |
- |
- |
PREDICTED: uncharacterized protein LOC105646742 [Jatropha curcas] |
| 11 |
Hb_009680_040 |
0.277357288 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_001449_100 |
0.2776296245 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 13 |
Hb_000230_150 |
0.2800089754 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_028271_020 |
0.2877756719 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 15 |
Hb_000154_040 |
0.2883901075 |
- |
- |
hypothetical protein JCGZ_25602 [Jatropha curcas] |
| 16 |
Hb_000331_640 |
0.2906593148 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_001789_050 |
0.2917825047 |
- |
- |
60S ribosomal protein L36-2 [Morus notabilis] |
| 18 |
Hb_002235_080 |
0.2941854952 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 19 |
Hb_000788_010 |
0.2959709301 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 20 |
Hb_002686_180 |
0.2963018242 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |