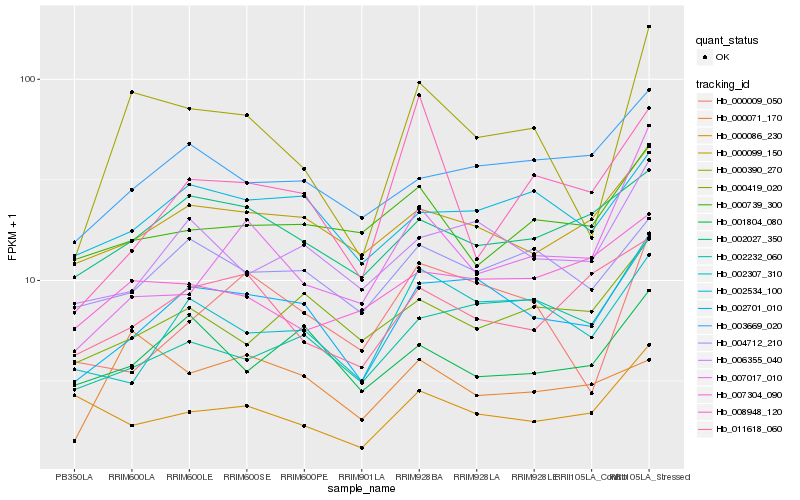
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_270 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002701_010 |
0.176885057 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000009_050 |
0.203027381 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 4 |
Hb_007017_010 |
0.2040450018 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 5 |
Hb_004712_210 |
0.210423774 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM50-like [Jatropha curcas] |
| 6 |
Hb_002307_310 |
0.2140288785 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 7 |
Hb_006355_040 |
0.2141021648 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_002534_100 |
0.2152973157 |
- |
- |
PREDICTED: malate dehydrogenase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003669_020 |
0.2153024928 |
- |
- |
PREDICTED: high mobility group nucleosome-binding domain-containing protein 5 [Populus euphratica] |
| 10 |
Hb_000099_150 |
0.2153868927 |
- |
- |
PREDICTED: calcium-dependent protein kinase 3 [Jatropha curcas] |
| 11 |
Hb_000071_170 |
0.2182746985 |
- |
- |
PREDICTED: uncharacterized protein LOC105628261 [Jatropha curcas] |
| 12 |
Hb_000739_300 |
0.2203480646 |
- |
- |
PREDICTED: uncharacterized protein LOC105643092 [Jatropha curcas] |
| 13 |
Hb_002027_350 |
0.2203626712 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 14 |
Hb_008948_120 |
0.2246095336 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_002232_060 |
0.2252375135 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 16 |
Hb_007304_090 |
0.2258206664 |
- |
- |
PREDICTED: uncharacterized protein LOC105645120 [Jatropha curcas] |
| 17 |
Hb_001804_080 |
0.2258409305 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 18 |
Hb_011618_060 |
0.2267159064 |
- |
- |
PREDICTED: DNA-repair protein XRCC1 [Jatropha curcas] |
| 19 |
Hb_000419_020 |
0.2278890608 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000086_230 |
0.2281425389 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |