| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
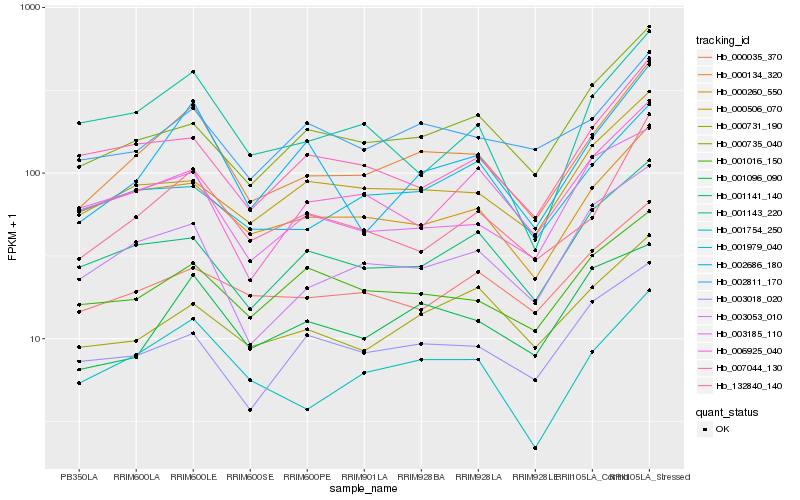
Hb_000134_320 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa013450mg [Prunus persica] |
| 2 |
Hb_132840_140 |
0.0918768791 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1 [Vitis vinifera] |
| 3 |
Hb_001754_250 |
0.0952040644 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 4 |
Hb_002686_180 |
0.1035760888 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 5 |
Hb_003053_010 |
0.1065698174 |
- |
- |
PREDICTED: uncharacterized protein LOC105641481 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000260_550 |
0.1081110073 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 7 |
Hb_002811_170 |
0.1122960117 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 8 |
Hb_001096_090 |
0.1143664431 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001141_140 |
0.1149602589 |
- |
- |
20S proteasome beta subunit D3 [Hevea brasiliensis] |
| 10 |
Hb_000506_070 |
0.1159245608 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 11 |
Hb_000735_040 |
0.1173973116 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 12 |
Hb_000731_190 |
0.1191820627 |
- |
- |
PREDICTED: cardiolipin synthase (CMP-forming), mitochondrial [Jatropha curcas] |
| 13 |
Hb_000035_370 |
0.1196303826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007044_130 |
0.1217989644 |
- |
- |
PREDICTED: cyclin-B1-2 [Jatropha curcas] |
| 15 |
Hb_006925_040 |
0.1219233541 |
- |
- |
60S ribosomal protein L24-1 isoform 2 [Theobroma cacao] |
| 16 |
Hb_001016_150 |
0.122478233 |
- |
- |
Rop1 [Hevea brasiliensis] |
| 17 |
Hb_001979_040 |
0.1229367694 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1-like [Gossypium raimondii] |
| 18 |
Hb_001143_220 |
0.1247276286 |
- |
- |
20S proteasome beta subunit C2 [Hevea brasiliensis] |
| 19 |
Hb_003185_110 |
0.1254054387 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 7-2 [Nelumbo nucifera] |
| 20 |
Hb_003018_020 |
0.1275467738 |
- |
- |
PREDICTED: uncharacterized protein LOC105628697 [Jatropha curcas] |