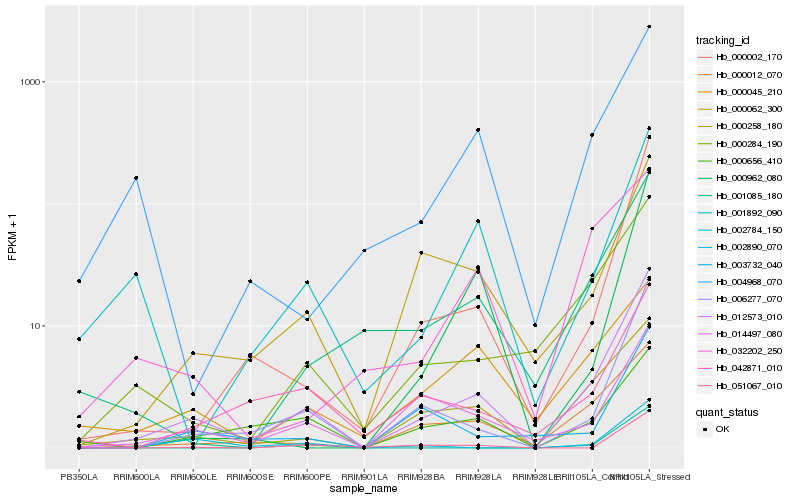
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012573_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 2 |
Hb_006277_070 |
0.1542443839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000002_170 |
0.1783584288 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B [Jatropha curcas] |
| 4 |
Hb_000258_180 |
0.1807148985 |
- |
- |
PREDICTED: glutathione S-transferase U10-like [Vitis vinifera] |
| 5 |
Hb_000012_070 |
0.18266146 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_000284_190 |
0.1961311757 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 7 |
Hb_000062_300 |
0.1993808773 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 8 |
Hb_001085_180 |
0.2089069965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_014497_080 |
0.2121449083 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 10 |
Hb_042871_010 |
0.2127939605 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 11 |
Hb_002784_150 |
0.2224941132 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 12 |
Hb_004968_070 |
0.223665998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003732_040 |
0.2263436407 |
- |
- |
putative esterase family protein [Populus trichocarpa] |
| 14 |
Hb_002890_070 |
0.2279818241 |
- |
- |
ATP-dependent RNA helicase family protein [Populus trichocarpa] |
| 15 |
Hb_051067_010 |
0.2290858596 |
- |
- |
- |
| 16 |
Hb_000656_410 |
0.2327357058 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 17 |
Hb_000962_080 |
0.2353456386 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 18 |
Hb_001892_090 |
0.2374937155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000045_210 |
0.2432443501 |
- |
- |
- |
| 20 |
Hb_032202_250 |
0.2471779348 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |