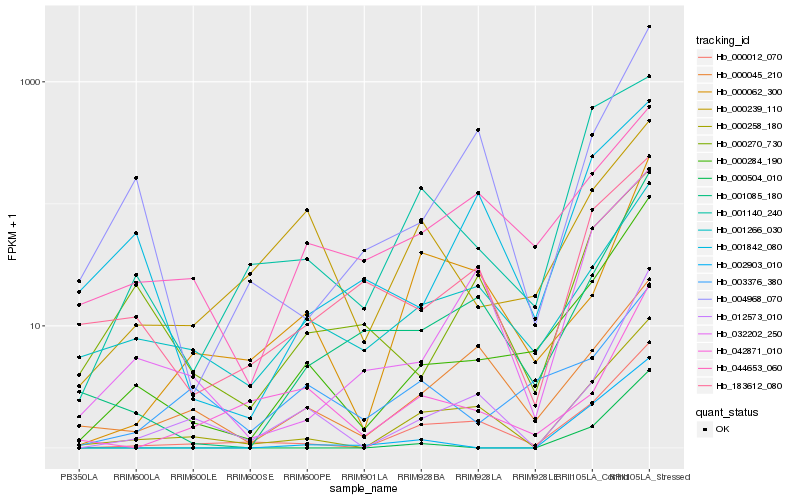
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000284_190 |
0.0 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 2 |
Hb_001085_180 |
0.1808049028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000012_070 |
0.1830770555 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_012573_010 |
0.1961311757 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 5 |
Hb_000258_180 |
0.1984733082 |
- |
- |
PREDICTED: glutathione S-transferase U10-like [Vitis vinifera] |
| 6 |
Hb_001266_030 |
0.2113738687 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_032202_250 |
0.2126085046 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 8 |
Hb_002903_010 |
0.2225071682 |
- |
- |
60S ribosomal protein L37-3 [Morus notabilis] |
| 9 |
Hb_044653_060 |
0.2229951556 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 10 |
Hb_004968_070 |
0.2247562363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001140_240 |
0.2275901272 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_000270_730 |
0.2296707569 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 13 |
Hb_001842_080 |
0.2321815185 |
- |
- |
- |
| 14 |
Hb_000045_210 |
0.2344944245 |
- |
- |
- |
| 15 |
Hb_000239_110 |
0.2358055687 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_003376_380 |
0.2419296879 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 17 |
Hb_000504_010 |
0.2421027563 |
- |
- |
oleosin 18.2 kDa-like [Jatropha curcas] |
| 18 |
Hb_183612_080 |
0.2452784043 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 19 |
Hb_042871_010 |
0.2511688846 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 20 |
Hb_000062_300 |
0.2513485961 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |