| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
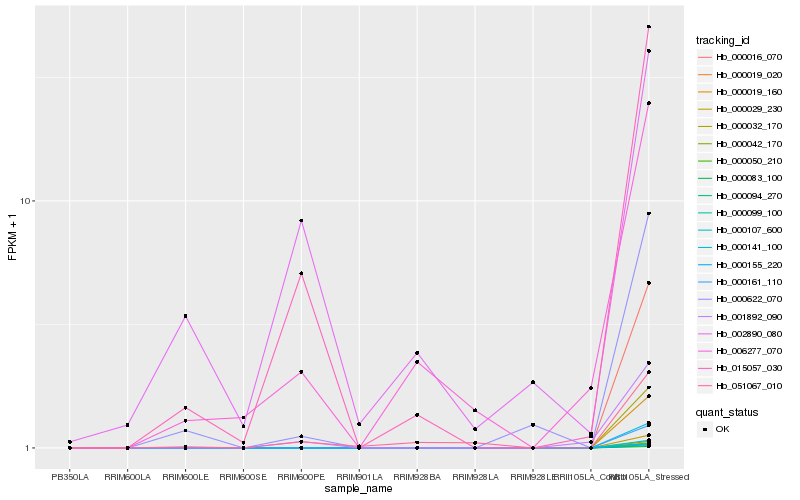
Hb_015057_030 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52, partial [Jatropha curcas] |
| 2 |
Hb_001892_090 |
0.1634079181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006277_070 |
0.1926863604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000622_070 |
0.1950213607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_051067_010 |
0.2040193766 |
- |
- |
- |
| 6 |
Hb_002890_080 |
0.2091292638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000016_070 |
0.2186860816 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box protein SOC1-like, partial [Nicotiana sylvestris] |
| 8 |
Hb_000019_020 |
0.2186860816 |
- |
- |
hypothetical protein POPTR_0017s08820g, partial [Populus trichocarpa] |
| 9 |
Hb_000019_160 |
0.2186860816 |
- |
- |
PREDICTED: RING finger protein 10 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000029_230 |
0.2186860816 |
- |
- |
hypothetical protein ARALYDRAFT_496947 [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_000032_170 |
0.2186860816 |
- |
- |
- |
| 12 |
Hb_000042_170 |
0.2186860816 |
transcription factor |
TF Family: ULT |
Protein ULTRAPETALA2, putative [Ricinus communis] |
| 13 |
Hb_000050_210 |
0.2186860816 |
- |
- |
PREDICTED: uncharacterized protein LOC105797515 [Gossypium raimondii] |
| 14 |
Hb_000083_100 |
0.2186860816 |
- |
- |
- |
| 15 |
Hb_000094_270 |
0.2186860816 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000099_100 |
0.2186860816 |
- |
- |
- |
| 17 |
Hb_000107_600 |
0.2186860816 |
- |
- |
- |
| 18 |
Hb_000141_100 |
0.2186860816 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Jatropha curcas] |
| 19 |
Hb_000155_220 |
0.2186860816 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 9-like [Jatropha curcas] |
| 20 |
Hb_000161_110 |
0.2186860816 |
- |
- |
hypothetical protein POPTR_0012s02100g [Populus trichocarpa] |