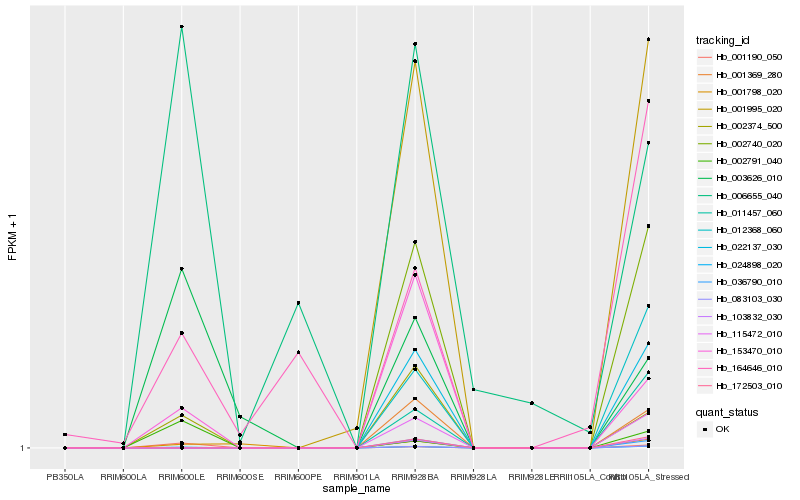
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024898_020 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_002374_500 |
0.2188596354 |
- |
- |
PREDICTED: putative F-box protein PP2-B12 [Gossypium raimondii] |
| 3 |
Hb_153470_010 |
0.2799694575 |
- |
- |
PREDICTED: uncharacterized protein LOC104804963 [Tarenaya hassleriana] |
| 4 |
Hb_001798_020 |
0.3036181053 |
- |
- |
PREDICTED: uncharacterized protein LOC104594032 [Nelumbo nucifera] |
| 5 |
Hb_164646_010 |
0.3076726697 |
- |
- |
PREDICTED: acidic endochitinase-like [Pyrus x bretschneideri] |
| 6 |
Hb_083103_030 |
0.3249518485 |
- |
- |
putative retrotransposon protein [Malus domestica] |
| 7 |
Hb_103832_030 |
0.3249730275 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 8 |
Hb_172503_010 |
0.3249825008 |
- |
- |
PREDICTED: uncharacterized protein LOC105647154 [Jatropha curcas] |
| 9 |
Hb_002791_040 |
0.3254020836 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 10 |
Hb_115472_010 |
0.3254876073 |
- |
- |
- |
| 11 |
Hb_002740_020 |
0.3257804935 |
- |
- |
- |
| 12 |
Hb_022137_030 |
0.3264619023 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 13 |
Hb_006655_040 |
0.3277134508 |
- |
- |
tau class glutathione transferase GSTU15 [Populus trichocarpa] |
| 14 |
Hb_001190_050 |
0.3301628204 |
- |
- |
hypothetical protein JCGZ_20709 [Jatropha curcas] |
| 15 |
Hb_001995_020 |
0.3312985585 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 16 |
Hb_003626_010 |
0.3337391946 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 17 |
Hb_036790_010 |
0.3355640752 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 18 |
Hb_012368_060 |
0.3404697125 |
- |
- |
- |
| 19 |
Hb_011457_060 |
0.340586611 |
- |
- |
PREDICTED: uncharacterized protein LOC105649957 [Jatropha curcas] |
| 20 |
Hb_001369_280 |
0.3408134955 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g64320, mitochondrial-like isoform X3 [Solanum tuberosum] |