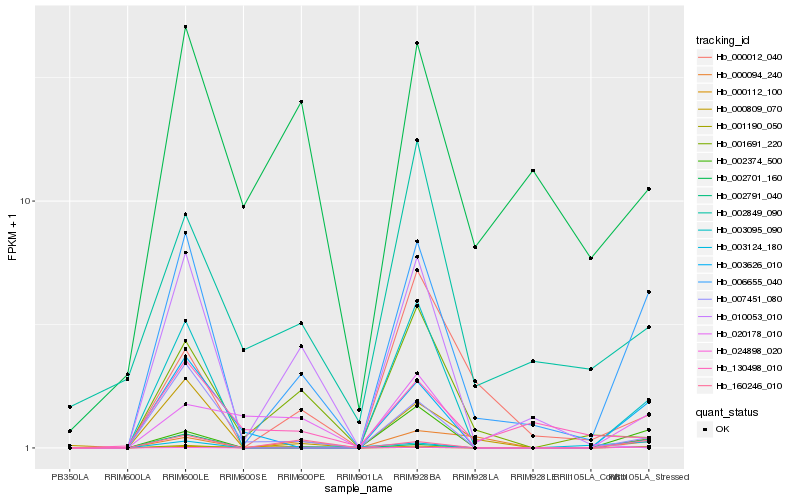
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006655_040 |
0.0 |
- |
- |
tau class glutathione transferase GSTU15 [Populus trichocarpa] |
| 2 |
Hb_000809_070 |
0.2594691733 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 36-like [Jatropha curcas] |
| 3 |
Hb_003095_090 |
0.2667162927 |
- |
- |
hypothetical protein JCGZ_10029 [Jatropha curcas] |
| 4 |
Hb_003626_010 |
0.2740336703 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 5 |
Hb_160246_010 |
0.2785248951 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_007451_080 |
0.2905915159 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 7 |
Hb_002374_500 |
0.2997276727 |
- |
- |
PREDICTED: putative F-box protein PP2-B12 [Gossypium raimondii] |
| 8 |
Hb_002701_160 |
0.3144420099 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 9 |
Hb_001691_220 |
0.3160997608 |
- |
- |
PREDICTED: uncharacterized protein LOC105649537 [Jatropha curcas] |
| 10 |
Hb_002849_090 |
0.3162871245 |
- |
- |
hypothetical protein POPTR_0002s16380g [Populus trichocarpa] |
| 11 |
Hb_000012_040 |
0.3175951806 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 12 |
Hb_003124_180 |
0.3234774366 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_002791_040 |
0.3250722726 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 14 |
Hb_024898_020 |
0.3277134508 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_001190_050 |
0.3287686404 |
- |
- |
hypothetical protein JCGZ_20709 [Jatropha curcas] |
| 16 |
Hb_130498_010 |
0.3347427868 |
- |
- |
Laccase 17 [Theobroma cacao] |
| 17 |
Hb_000094_240 |
0.3355168067 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 18 |
Hb_000112_100 |
0.3355885074 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 19 |
Hb_020178_010 |
0.3372904925 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 20 |
Hb_010053_010 |
0.3387254049 |
- |
- |
hypothetical protein POPTR_0515s00220g [Populus trichocarpa] |