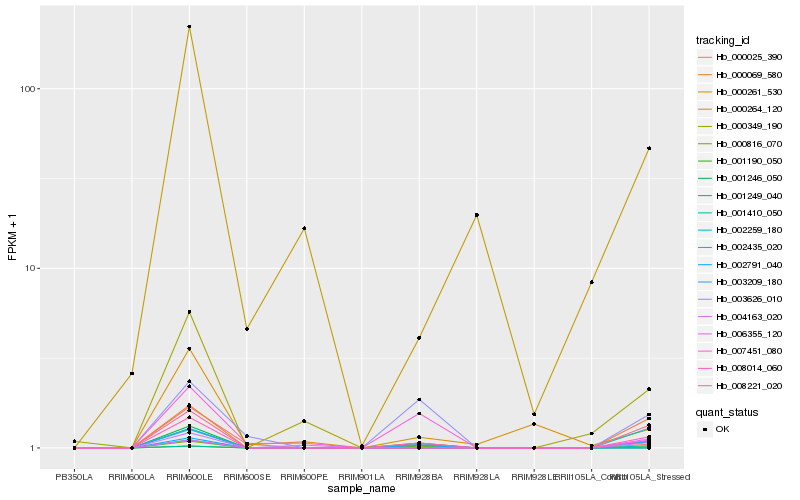
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008014_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001190_050 |
0.1242907832 |
- |
- |
hypothetical protein JCGZ_20709 [Jatropha curcas] |
| 3 |
Hb_002791_040 |
0.1276570638 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 4 |
Hb_001246_050 |
0.2205078868 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 5 |
Hb_008221_020 |
0.2422906375 |
- |
- |
hypothetical protein JCGZ_15014 [Jatropha curcas] |
| 6 |
Hb_000069_580 |
0.2553193062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000025_390 |
0.2757249904 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 8 |
Hb_002435_020 |
0.276287709 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 9 |
Hb_000349_190 |
0.2893634932 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 10 |
Hb_003626_010 |
0.2907806041 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 11 |
Hb_002259_180 |
0.3166331998 |
- |
- |
- |
| 12 |
Hb_004163_020 |
0.321304461 |
- |
- |
- |
| 13 |
Hb_007451_080 |
0.3278776089 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 14 |
Hb_006355_120 |
0.3293779735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000264_120 |
0.3302164442 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 16 |
Hb_001410_050 |
0.3329672042 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 17 |
Hb_000261_530 |
0.3357066513 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 18 |
Hb_003209_180 |
0.3459439083 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme [Morus notabilis] |
| 19 |
Hb_001249_040 |
0.3509374083 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 20 |
Hb_000816_070 |
0.3543407983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |