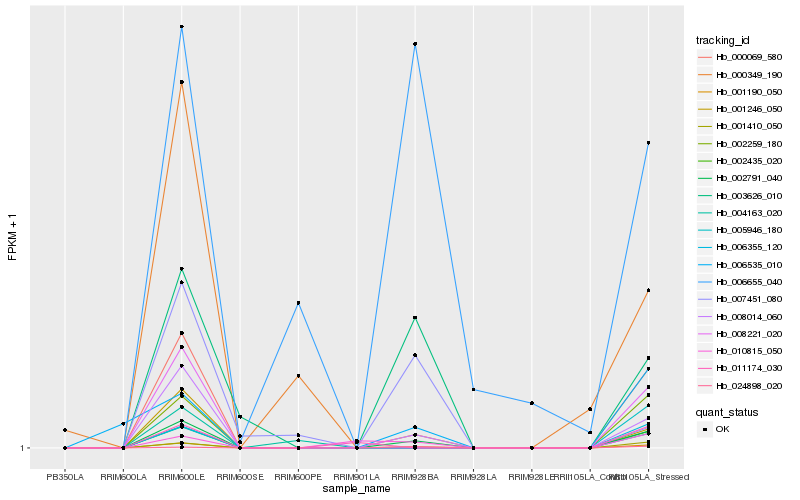
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001190_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_20709 [Jatropha curcas] |
| 2 |
Hb_002791_040 |
0.0057263167 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 3 |
Hb_008014_060 |
0.1242907832 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000069_580 |
0.2604507408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008221_020 |
0.260758661 |
- |
- |
hypothetical protein JCGZ_15014 [Jatropha curcas] |
| 6 |
Hb_003626_010 |
0.2622447034 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 7 |
Hb_002435_020 |
0.2647287399 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 8 |
Hb_002259_180 |
0.2823109994 |
- |
- |
- |
| 9 |
Hb_006355_120 |
0.2895591987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001410_050 |
0.2917135107 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 11 |
Hb_001246_050 |
0.2918821672 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 12 |
Hb_006535_010 |
0.3128979313 |
- |
- |
- |
| 13 |
Hb_006655_040 |
0.3287686404 |
- |
- |
tau class glutathione transferase GSTU15 [Populus trichocarpa] |
| 14 |
Hb_024898_020 |
0.3301628204 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_010815_050 |
0.3402910451 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 16 |
Hb_000349_190 |
0.3505542733 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 17 |
Hb_011174_030 |
0.3524317268 |
- |
- |
- |
| 18 |
Hb_007451_080 |
0.3628935035 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 19 |
Hb_004163_020 |
0.3639030594 |
- |
- |
- |
| 20 |
Hb_005946_180 |
0.3640231803 |
- |
- |
PREDICTED: uncharacterized protein LOC100818788 [Glycine max] |