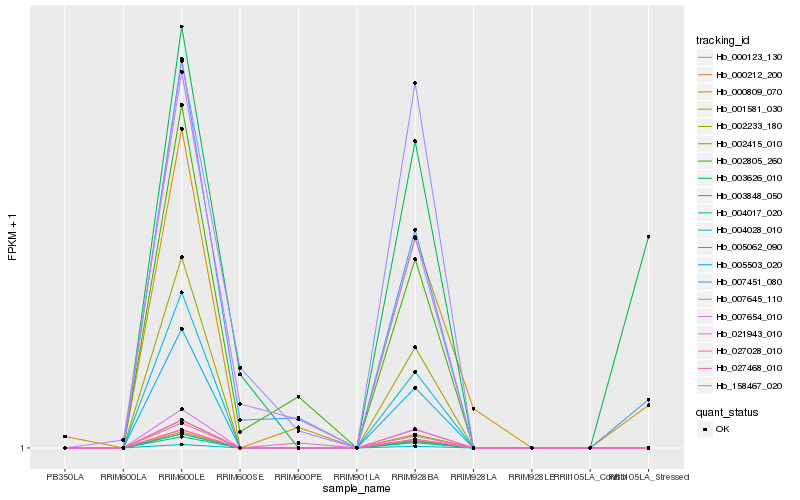
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007451_080 |
0.0 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 2 |
Hb_002805_260 |
0.1775493478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007654_010 |
0.1804140992 |
- |
- |
valacyclovir hydrolase, putative [Ricinus communis] |
| 4 |
Hb_000809_070 |
0.2204849719 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 36-like [Jatropha curcas] |
| 5 |
Hb_007645_110 |
0.2303253017 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 6 |
Hb_003626_010 |
0.2322559867 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 7 |
Hb_027028_010 |
0.2402819821 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 8 |
Hb_021943_010 |
0.2427643038 |
- |
- |
- |
| 9 |
Hb_004017_020 |
0.2427691779 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 10 |
Hb_027468_010 |
0.2427733944 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_003848_050 |
0.2427740262 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 12 |
Hb_001581_030 |
0.2427760032 |
- |
- |
PREDICTED: putative F-box/FBD/LRR-repeat protein At5g56810 [Jatropha curcas] |
| 13 |
Hb_000212_200 |
0.2427781209 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 14 |
Hb_004028_010 |
0.2427938855 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_158467_020 |
0.2428020387 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 16 |
Hb_002415_010 |
0.2428106783 |
- |
- |
PREDICTED: uncharacterized protein LOC105775231 [Gossypium raimondii] |
| 17 |
Hb_005503_020 |
0.2428163556 |
- |
- |
Hypothetical protein glysoja_025074 [Glycine soja] |
| 18 |
Hb_005062_090 |
0.2428275855 |
- |
- |
ferredoxin [Vibrio natriegens] |
| 19 |
Hb_002233_180 |
0.2428582253 |
- |
- |
- |
| 20 |
Hb_000123_130 |
0.2429247981 |
- |
- |
hypothetical protein POPTR_0008s05200g, partial [Populus trichocarpa] |