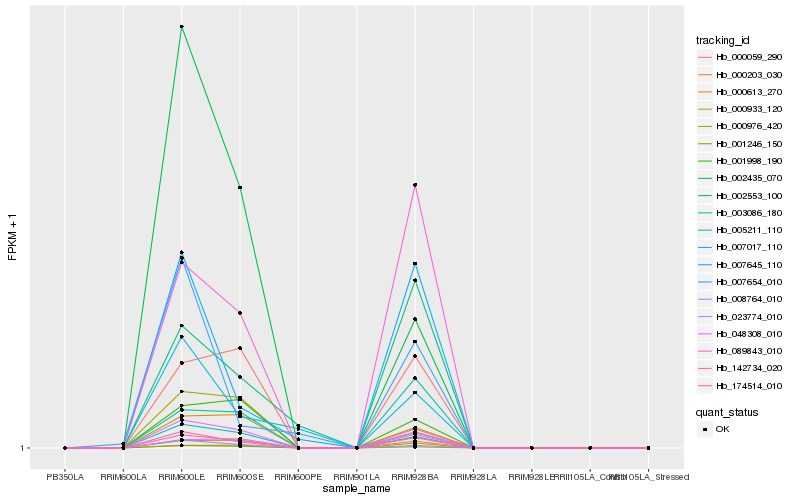
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142734_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X1 [Gossypium raimondii] |
| 2 |
Hb_048308_010 |
0.1302333692 |
- |
- |
Peroxidase superfamily protein [Theobroma cacao] |
| 3 |
Hb_005211_110 |
0.1311121609 |
- |
- |
hypothetical protein POPTR_0010s13790g [Populus trichocarpa] |
| 4 |
Hb_008764_010 |
0.1372641925 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001246_150 |
0.1385565997 |
- |
- |
hypothetical protein MIMGU_mgv1a021562mg [Erythranthe guttata] |
| 6 |
Hb_174514_010 |
0.1659763662 |
- |
- |
flavin-containing monooxygenase family protein [Populus trichocarpa] |
| 7 |
Hb_007645_110 |
0.1805984816 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 8 |
Hb_000203_030 |
0.1841372724 |
- |
- |
- |
| 9 |
Hb_089843_010 |
0.1928379608 |
- |
- |
hypothetical protein JCGZ_05278 [Jatropha curcas] |
| 10 |
Hb_000059_290 |
0.2078492299 |
- |
- |
pterin-4-alpha-carbinolamine dehydratase, putative [Ricinus communis] |
| 11 |
Hb_007017_110 |
0.2123114956 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 12 |
Hb_001998_190 |
0.2207979041 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |
| 13 |
Hb_003086_180 |
0.2235352102 |
- |
- |
PREDICTED: RING-H2 finger protein ATL70-like [Pyrus x bretschneideri] |
| 14 |
Hb_007654_010 |
0.2283712108 |
- |
- |
valacyclovir hydrolase, putative [Ricinus communis] |
| 15 |
Hb_000933_120 |
0.2290426252 |
- |
- |
hypothetical protein L484_011925 [Morus notabilis] |
| 16 |
Hb_000976_420 |
0.2296191597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002553_100 |
0.2298073642 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 18 |
Hb_000613_270 |
0.2310697862 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 19 |
Hb_002435_070 |
0.2321875737 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 20 |
Hb_023774_010 |
0.2337998375 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |