| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
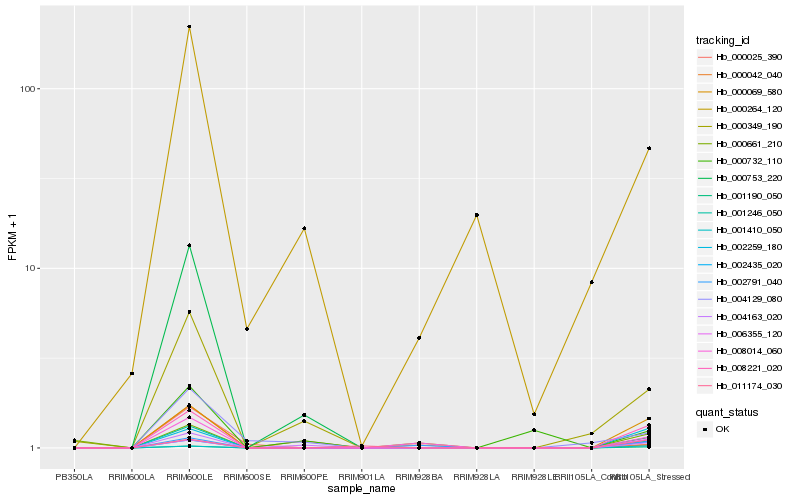
Hb_001246_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 2 |
Hb_008221_020 |
0.1194727775 |
- |
- |
hypothetical protein JCGZ_15014 [Jatropha curcas] |
| 3 |
Hb_000069_580 |
0.1482327999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002435_020 |
0.1867330365 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_008014_060 |
0.2205078868 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004163_020 |
0.2434802706 |
- |
- |
- |
| 7 |
Hb_002259_180 |
0.2487739664 |
- |
- |
- |
| 8 |
Hb_000349_190 |
0.2488244264 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 9 |
Hb_006355_120 |
0.2665915938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001410_050 |
0.2715042545 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 11 |
Hb_004129_080 |
0.2915359439 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 12 |
Hb_001190_050 |
0.2918821672 |
- |
- |
hypothetical protein JCGZ_20709 [Jatropha curcas] |
| 13 |
Hb_002791_040 |
0.2965918086 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 14 |
Hb_000264_120 |
0.296787554 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 15 |
Hb_000025_390 |
0.3038029472 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 16 |
Hb_000661_210 |
0.3125649137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000042_040 |
0.3187645582 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 18 |
Hb_011174_030 |
0.3208732114 |
- |
- |
- |
| 19 |
Hb_000732_110 |
0.3281071276 |
- |
- |
hypothetical protein JCGZ_21339 [Jatropha curcas] |
| 20 |
Hb_000753_220 |
0.3353997906 |
- |
- |
hypothetical protein RCOM_1467930 [Ricinus communis] |