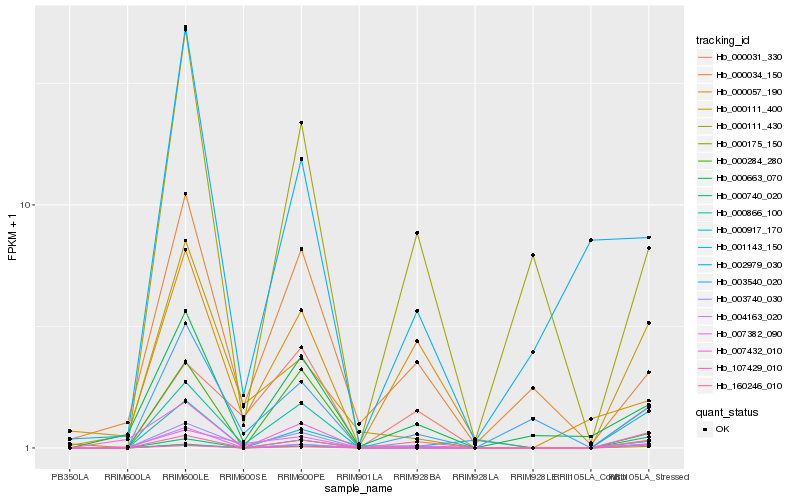
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_150 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein B456_009G311700 [Gossypium raimondii] |
| 2 |
Hb_160246_010 |
0.1501409747 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_001143_150 |
0.2009165674 |
- |
- |
PREDICTED: mitochondrial protein import protein MAS5-like [Populus euphratica] |
| 4 |
Hb_003740_030 |
0.2267327571 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 5 |
Hb_000663_070 |
0.2397756208 |
- |
- |
cyclin A, putative [Ricinus communis] |
| 6 |
Hb_000111_430 |
0.2412397878 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Jatropha curcas] |
| 7 |
Hb_004163_020 |
0.2517453668 |
- |
- |
- |
| 8 |
Hb_003540_020 |
0.264821859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000917_170 |
0.2738755977 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 10 |
Hb_000740_020 |
0.27827599 |
transcription factor |
TF Family: MYB-related |
asymmetric leaves1 and rough sheath, putative [Ricinus communis] |
| 11 |
Hb_007432_010 |
0.2841000003 |
- |
- |
hypothetical protein B456_004G088300 [Gossypium raimondii] |
| 12 |
Hb_000057_190 |
0.2841033131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000866_100 |
0.2844579479 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_107429_010 |
0.2846295062 |
- |
- |
PREDICTED: calcium-dependent protein kinase 34 [Jatropha curcas] |
| 15 |
Hb_000034_150 |
0.2850095844 |
- |
- |
PREDICTED: kinesin-like protein BC2 [Jatropha curcas] |
| 16 |
Hb_000111_400 |
0.2855000065 |
- |
- |
hypothetical protein POPTR_0003s19310g [Populus trichocarpa] |
| 17 |
Hb_007382_090 |
0.2856038506 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_000031_330 |
0.2859551973 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0157s00200g, partial [Populus trichocarpa] |
| 19 |
Hb_002979_030 |
0.2930883165 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000284_280 |
0.297850145 |
- |
- |
- |