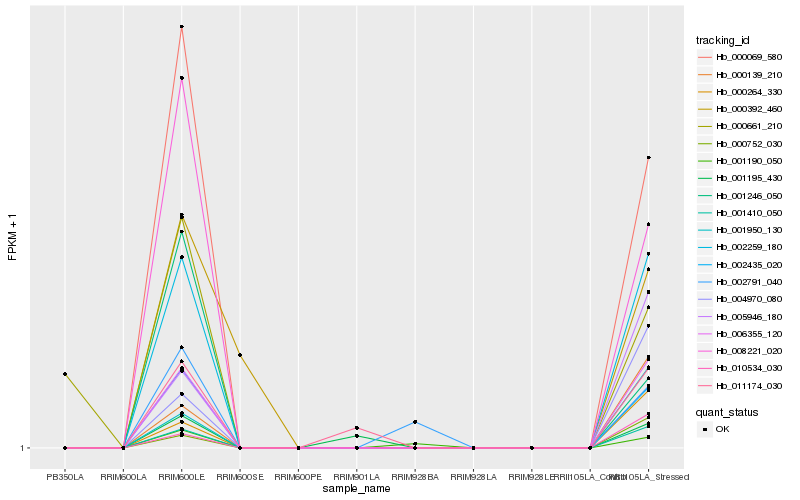
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_006355_120 |
0.0182627094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001410_050 |
0.0233072809 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 4 |
Hb_002435_020 |
0.0631751112 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_000069_580 |
0.1020391055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008221_020 |
0.1308878915 |
- |
- |
hypothetical protein JCGZ_15014 [Jatropha curcas] |
| 7 |
Hb_005946_180 |
0.1517703744 |
- |
- |
PREDICTED: uncharacterized protein LOC100818788 [Glycine max] |
| 8 |
Hb_000139_210 |
0.1602972349 |
- |
- |
- |
| 9 |
Hb_000264_330 |
0.1629053933 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 10 |
Hb_004970_080 |
0.1717137813 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 11 |
Hb_001950_130 |
0.1740571598 |
- |
- |
- |
| 12 |
Hb_010534_030 |
0.1752963209 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 13 |
Hb_000752_030 |
0.1757071601 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_0978110 [Ricinus communis] |
| 14 |
Hb_011174_030 |
0.2343385351 |
- |
- |
- |
| 15 |
Hb_001246_050 |
0.2487739664 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 16 |
Hb_001190_050 |
0.2823109994 |
- |
- |
hypothetical protein JCGZ_20709 [Jatropha curcas] |
| 17 |
Hb_002791_040 |
0.2860100093 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 18 |
Hb_000661_210 |
0.3095038979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000392_460 |
0.3103935949 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 20 |
Hb_001195_430 |
0.3110303402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |